Method for quickly detecting proteins in bombyx batryticatus according to biological mass spectrometry technology
A biological mass spectrometry and protein technology, applied in the field of traditional Chinese medicine, can solve the problems of inaccurate determination of protein molecular weight, cumbersome steps, time and reagent consumption, etc., and achieve the effect of rich mass-to-charge ratio signals, fast measurement speed, and shortened sample preparation time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
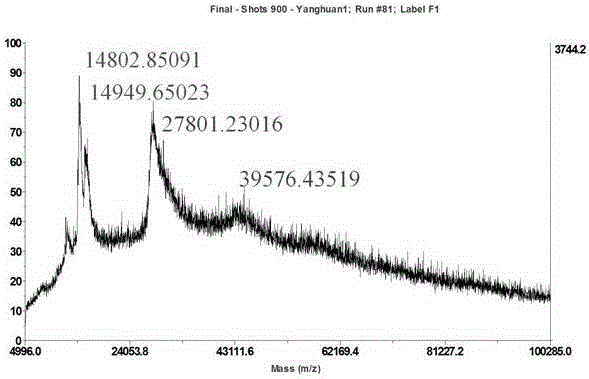
[0038] Crush 20 mg of silkworms to obtain fine powder, add 200 μL of water, ultrasonically extract for 30 minutes, and centrifuge at 12,000 r / min for 15 minutes; slowly add the supernatant to 6 mL of saturated ammonium sulfate solution, vortex for a few seconds, and stand overnight at 4°C; The mixed solution was centrifuged again at 12000r / min for 15 minutes, the supernatant was discarded, and the precipitate was collected; the above precipitate was dissolved with about 100 μL of water to obtain a solution; the protein in the sample solution was adsorbed by reversed-phase bonded solid-phase extraction MilliporeC4Ziptip at 70 % Acetonitrile 10 μL eluted to obtain the test solution; draw 1 μL of the test solution, apply the sample, then take 1 μL of the saturated 3,5-dimethoxy-4-hydroxycinnamic acid matrix solution, mix and evaporate to dryness; the sample Plates were loaded into a MALDI-TOF mass spectrometer for analysis.
Embodiment 2
[0040] Crush 20 mg of silkworms to obtain fine powder, add 1000 μL of water, ultrasonically extract for 30 minutes, and centrifuge at 12000 r / min for 15 minutes; slowly add the supernatant to 50 mL of saturated ammonium sulfate solution, vortex for a few seconds, and stand overnight at 4 °C; The mixed solution was centrifuged again at 12000r / min for 15 minutes, the supernatant was discarded, and the precipitate was collected; the above precipitate was dissolved in about 100 μL of water to obtain a solution; the protein in the sample solution was adsorbed by reverse-phase bonded solid-phase extraction MilliporeC4Ziptip, and the protein in the sample solution was extracted at 80 % Acetonitrile 5 μL eluted to obtain the test solution; draw 0.5 μL of the test solution, apply the sample, then take 0.5 μL of the saturated 3,5-dimethoxy-4-hydroxycinnamic acid matrix solution, mix, and evaporate to dryness; Load the sample plate into a MALDI-TOF mass spectrometer for analysis.
Embodiment 3
[0042] Crush 20 mg of silkworms to obtain a fine powder, add 1200 μL of water, ultrasonically extract for 30 minutes, and centrifuge at 12,000 r / min for 15 minutes; slowly add the supernatant to 60 mL of saturated ammonium sulfate solution, vortex for a few seconds, and stand overnight at 4°C; The mixed solution was centrifuged again at 12000r / min for 15 minutes, the supernatant was discarded, and the precipitate was collected; the above precipitate was dissolved in about 100 μL of water to obtain a solution; the protein in the sample solution was adsorbed by reversed-phase bonded solid-phase extraction MilliporeC4Ziptip at 90 % Acetonitrile 5 μL eluted to obtain the test solution; draw 0.5 μL of the test solution, apply the sample, then take 0.5 μL of the saturated 3,5-dimethoxy-4-hydroxycinnamic acid matrix solution, mix, and evaporate to dryness; Load the sample plate into a MALDI-TOF mass spectrometer for analysis.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com