Detection primer set for Rickettsia beinii and its detection reagent, real-time fluorescent quantitative PCR method
A technology of real-time fluorescence quantification of Rickettsia bainierii, applied in the direction of recombinant DNA technology, DNA / RNA fragments, etc., can solve the problem of difficult early diagnosis of specific antibody detection results, time-consuming isolation of Rickettsia bainiette It is used to detect problems such as samples to achieve the effect of good gradient repeatability, stable repeatability and small error
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0035] The preparation method of Rickettsia bainiferi standard substance is:
[0036] First, the target gene was synthesized according to the Rickettsia beinii IS111a gene sequence published by NCBI (see SEQID NO: 4 for the sequence, synthesized by Shanghai Bioengineering Co., Ltd.) and the PMD18T vector (TAKARA Company) was ligated and transferred into E. coli, and directly extracted Plasmid DNA;
[0037] Plasmids were used as Rickettsia bainiferi standards for experiments. The plasmid had a 260nm / 280nm ratio of 2.0 and a concentration of 56.00 ng / μL.
[0038] Buffer ATL, Buffer AL, Buffer AW1, Buffer AW2, Buffer AE, and QIA&Mini spin column collection tubes used for extracting DNA from samples to be tested were from DNA extraction kits purchased from Beijing Quanshijin Biotechnology Co., Ltd.
Embodiment 1
[0040] The detection method of embodiment 1 Rickettsia bainiferi
[0041] Taking the nucleotide sequence shown in SEQ ID NO:1 as an upstream primer; using the nucleotide sequence shown in SEQ ID NO:2 as a downstream primer; using the nucleotide sequence shown in SEQ ID NO:3 as a probe to detect shellfish Rickettsiae.
[0042] According to the orthogonal verification of the matrix method, the optimal concentration and dosage of primers and probes were determined. Using the plasmid containing the Rickettsia bainiferi gene as a template, a fluorescent PCR reaction system was established, and the total reaction system was 25 μL. Under the condition that the template concentration is the same, use 5, 10, 15, 20 pM primer concentration and 5, 10, 20 pM probe concentration for primer and probe respectively, and use matrix method to screen the optimal concentration of primer and probe. Results The optimal primer concentration was 10pM, and the probe concentration was 10pM. The best...
Embodiment 2
[0051] Embodiment 2 specific detection
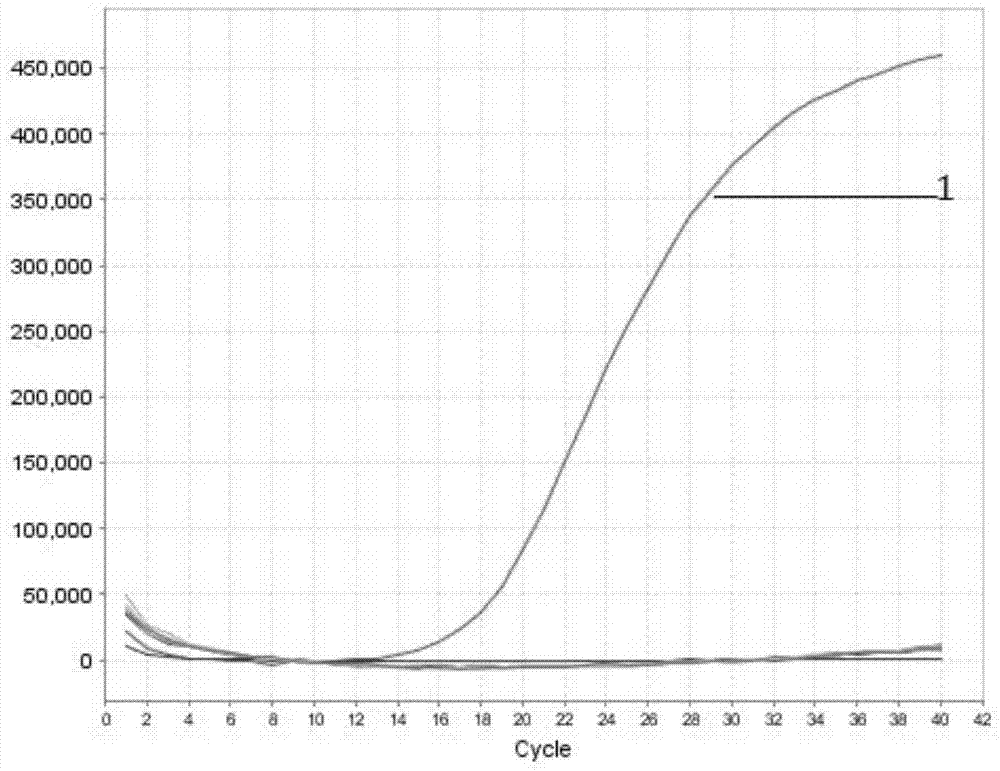
[0052] According to the reaction system and the conditions of Example 1, Rickettsia bainii, Rickettsia siberia, Yersinia pestis, Tularemia, Brucella, Escherichia coli, Listeria were detected respectively, and the results were as follows figure 1 . Through the amplification, only Rickettsia bainiferi showed an amplification curve, and the others did not have an S-shaped curve amplification, and the negative control was established, indicating good specificity. The results of the study showed that the established detection method had strong specificity and could be used for the specific identification of Rickettsia bainiferi.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com