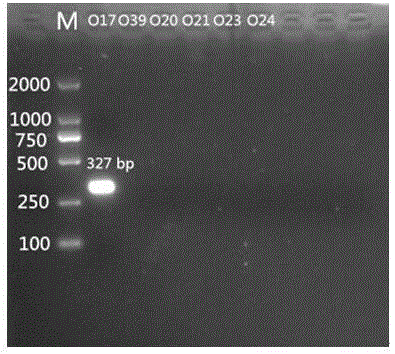
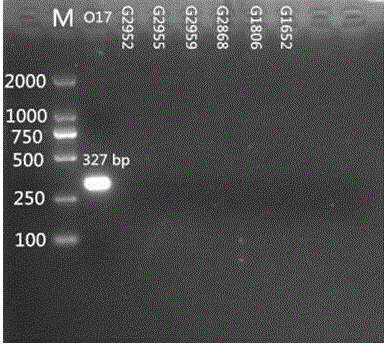
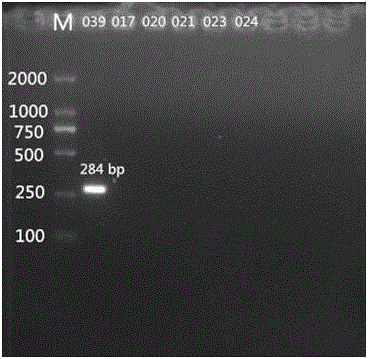
Specific nucleotides for citrobacter 017 and 039 and application of special nucleotides
A Citrobacter, nucleotide technology, applied in microorganism-based methods, biochemical equipment and methods, and microbial assay/inspection, etc., can solve the difficulty of sugar synthesis gene identification, complex polysaccharide antigen synthesis gene cluster composition, and genetic Unpredictable functions, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 : Genome Extraction
[0035] Citrobacter was cultured at 37°C in brain-heart infusion broth medium, the bacteria were collected, and the genome was extracted with a bacterial genome extraction kit (Kangwei Century CW0552). The specific steps are as follows:
[0036] 1. Take 1-5ml of bacterial culture (106-108 cells, no more than 2×109 cells), centrifuge at 10000rpm for 1 minute, and discard the supernatant.
[0037] 2. Add 180μl BufferGTL to the pellet and shake to resuspend the bacteria.
[0038] 3. Add 20 μl ProteinaseK, vortex to mix, and incubate at 56°C until the cells are completely lysed. During the incubation, invert or shake the centrifuge tube at regular intervals to disperse the sample.
[0039] 4. Add 4 μl of RNaseA solution (Product No. CW0601) with a concentration of 100 mg / ml, vortex to mix, incubate at room temperature for 5 minutes, and then proceed to step 5.
[0040] 5. Add 200μl BufferGL, vortex and mix well; add 200μl absolute ethano...
Embodiment 2
[0052] Example 2: sequence deciphering
[0053] The genomes of the standard strains of Citrobacter 017 and O39 serotypes were extracted, and the genomes of each serotype of Citrobacter were sequenced by Solexapair-end sequencing technology to obtain the sequence of the serotype, and the sequences were compared using Blast and PSI-Blast. The TMHMM2.0 program was used to predict the transmembrane structure, and the ClustalW program was used to perform sequence alignment and screen conservative and specific gene fragments. Finally, the O antigen gene cluster sequences and decipher results of each serotype of Citrobacter were obtained.
Embodiment 3
[0054] Example 3 : Primer design
[0055] According to the deciphering of gene clusters, we found that wzm and wzy The gene is indeed a serotype-specific gene, so a specific segment of the gene is selected to design specific primers.
[0056] Primer design is a core part of this invention. Primers were designed according to the specific genes described in the literature. wzm and wzy The gene is a relatively specific gene in the Citrobacter O antigen gene cluster, which can be used as the target gene for serotype identification. Import the above genes into PrimerPremier5 for primer design. The length of the primers should preferably be between 18 and 24 bp, and the Tm value should be around 55°C. A pair of primers are designed for each gene, with a single target band.
[0057] After the primers are designed, BLAST is performed in Genbank. The designed primers should not have too high sequence similarity with other related bacteria, so as to ensure that the primers ca...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com