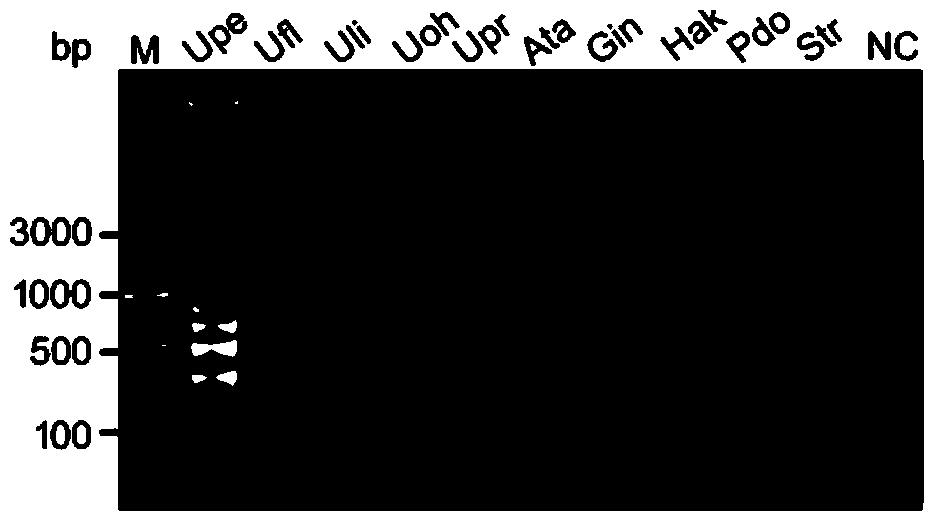Primer and probe sequences for Ulva pore lamp-lfd detection
A LAMP-LFD and probe sequence technology is applied in the field of primers and probe sequences for Ulva vulva LAMP-LFD detection to achieve clear and obvious detection results, personal and environmental safety, and obvious results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Establishment of a method for the detection of Ulva perforatum by LAMP-LFD technique
[0033] 1. Primer design: Design according to the coding sequence of the transcribed spacer in Ulva pore (GenBank accession number: HQ902008) published in NCBI, where the primer sequence is as follows:
[0034] UpeITS-F3: 5'-TAGCCTCACCTGAACTCAG-3'
[0035] UpeITS-B3: 5'-ACGGATATCTCGGCTCTC-3',
[0036] UpeITS-FIP: 5'-CGGCTGAAATACAGAGGCTCGTATCCTTCGTGGCTCACA-3',
[0037] UpeITS-BIP: 5'- GTTATTCCGACGCTGAGGCAGCAGAATTCCGTGAGTCAT-3',
[0038] UpeITS-LF: 5'-TAGGTAGCTCGCTACTCCTAC-3',
[0039] UpeITS-LB: 5'- GTGGTCTCATCCGAAGACTC-3', the 5' end of UpeITS-FIP is biotinylated;
[0040] Probe UpeITS-HP: 5'-GCAAGCGCGTGAGGGGTTAT-3', the 5' end is labeled with fluorescein isothiocyanate.
[0041] 2. Sample DNA extraction: According to the steps of Tissue Genomic DNA Extraction Kit (Spin Column Type) D3396-0, the genomic DNA of Ulva pore S66 isolate (Qiagen, Hilden, Germany) was extracted. The obt...
Embodiment 2
[0048] Specificity determination of Ulva pore LAMP-LFD detection using primers and probes of the invention
[0049] Using the designed specific primers and probes, respectively, to select Ulva pore S66, Enteromorpha spp. SDF12, Enteromorpha spp. Ulvaohnoi FJ4, Enteromorpha XS5 and other common Ulva green algae, as well as domestic common microalgae such as Alexandrium tamarina NMBjah048, annulus sp. Genomic DNA of algal species was used as a template, and LAMP-LFD detection was performed according to steps 3 and 4 of Example 1 above to verify the specificity of primers and probes, and double distilled water was used as a negative control. The result is as figure 1 and figure 2 As shown, using the electrophoresis method ( figure 1 ) and LFD ( figure 2 ) can only amplify the target bands from the genomic DNA samples of Ulva kongfu, and other samples have no amplified bands, indicating that the primers and probes provided by the present invention are used for LAMP-LFD d...
Embodiment 3
[0051] Sensitivity determination of Ulva pore LAMP-LFD detection using primers and probes of the present invention
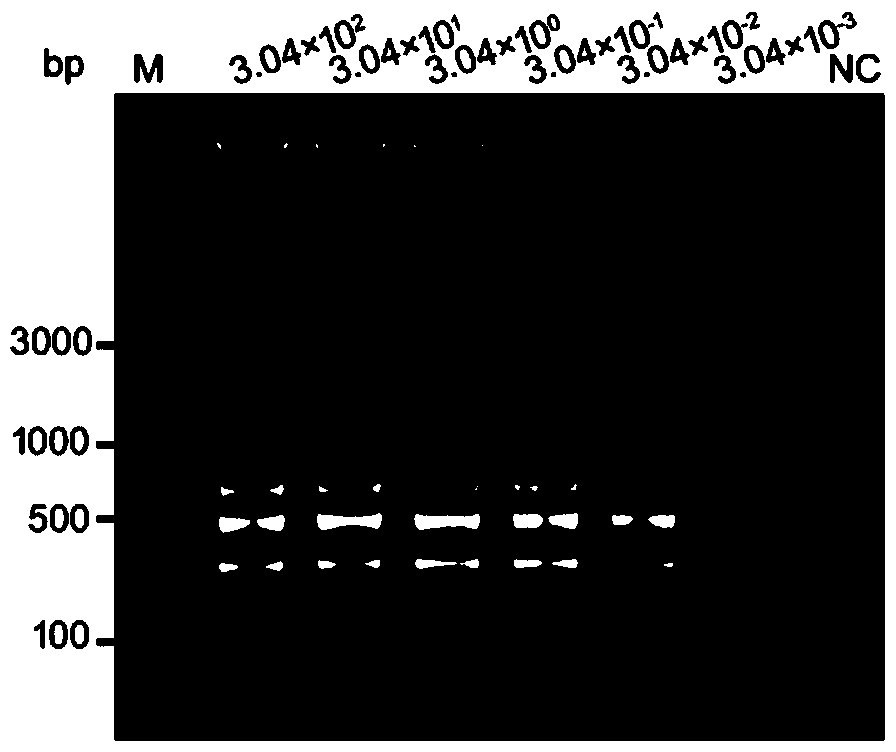
[0052] Adopt the method of step 2 of above-mentioned embodiment 1 to extract the genomic DNA of Ulva kongii, carry out 10 times gradient dilution, select 3.04 * 10 2 , 3.04×10 1 , 3.04×10 0 , 3.04×10 -1 , 3.04×10 -2 , 3.04×10 -3 pg / μL etc. were used as templates, and LAMP-LFD detection was performed according to steps 3 and 4 of Example 1 above to verify the sensitivity of primers and probes, and sterile deionized water was used as a negative control. The result is as image 3 , Figure 4 and Figure 5 As shown, the sensitivity of the LAMP-LFD detection using the primers and probes provided by the invention is 3.04×10 -2 pg / μL ( Figure 4 ), consistent with the sensitivity obtained by agarose gel electrophoresis detection of LAMP amplification products ( image 3 ), which is 1000 times that of the conventional PCR detection method established with Up...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com