Fluorescent quantitative PCR detection kit for clostridium difficile toxin A/B and detection method
A technology of Clostridium difficile toxin and Clostridium difficile, applied in the field of biomedicine, can solve the problems of difficult primer design, reduced quantity, cumbersome later verification, etc., and achieve reliable primer detection results and reliable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] 16SrDNA specific primers
[0056] The specific primer design method for Clostridium difficile 16S rDNA is as follows:
[0057] When the 16SrDNA sequences of different bacterial species are almost identical, the ability of 16SrDNA universal primers to identify bacterial species is limited. In order to solve this problem, we designed multiple primers, and after screening dozens of primers, we finally screened out 16SrDNA primers specific only to C.difficile, and identified C.difficile accordingly.
[0058] The most specific 16S rDNA primer sequences screened for detection of C.difficile are as follows:
[0059] C.diff-16s-F: TTGAGCGATTTACTTCGGTAAAGA
[0060] C.diff-16s-R:CCATCCTGTACTGGCTCACCT
[0061] 16SrDNA-specific primers for detection of C.difficile
[0062] Bacterial total genomic DNA was extracted from the feces of 234 clinical patients with intestinal diarrhea, and the most specific 16S rDNA primers screened for the detection of C.difficile were used to de...
Embodiment 2
[0069] Design method of specific primers for tcdA and tcdB gene quality control products, qPCR detection primers and probe sequences
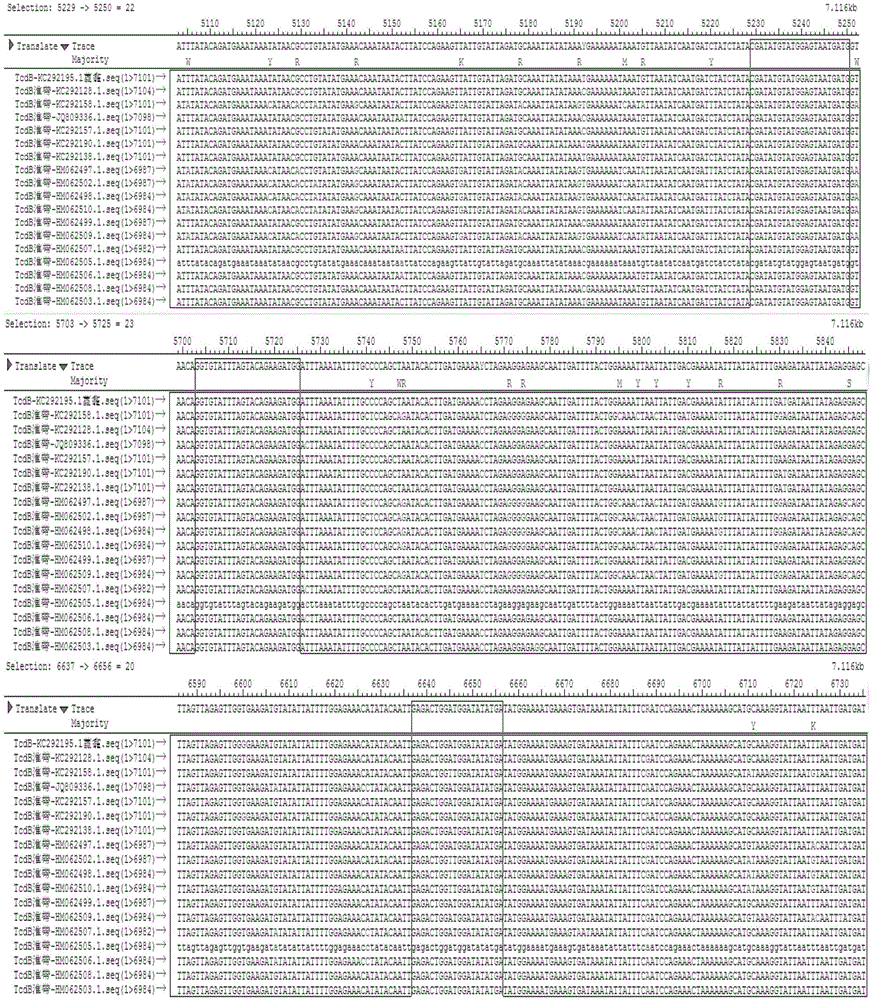
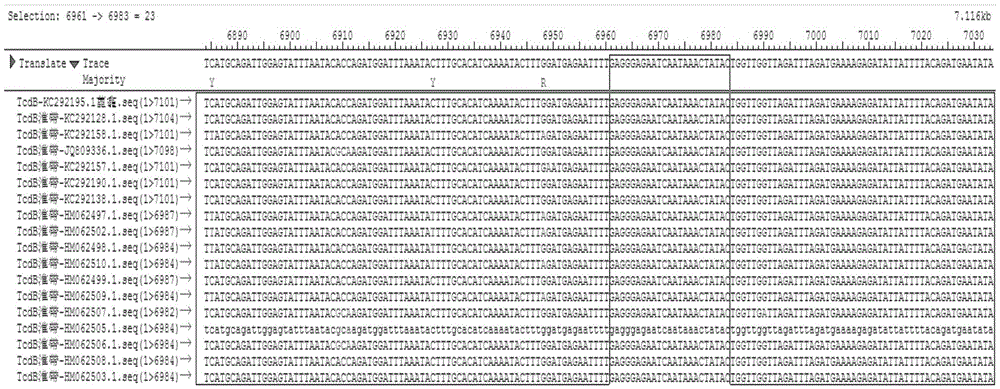
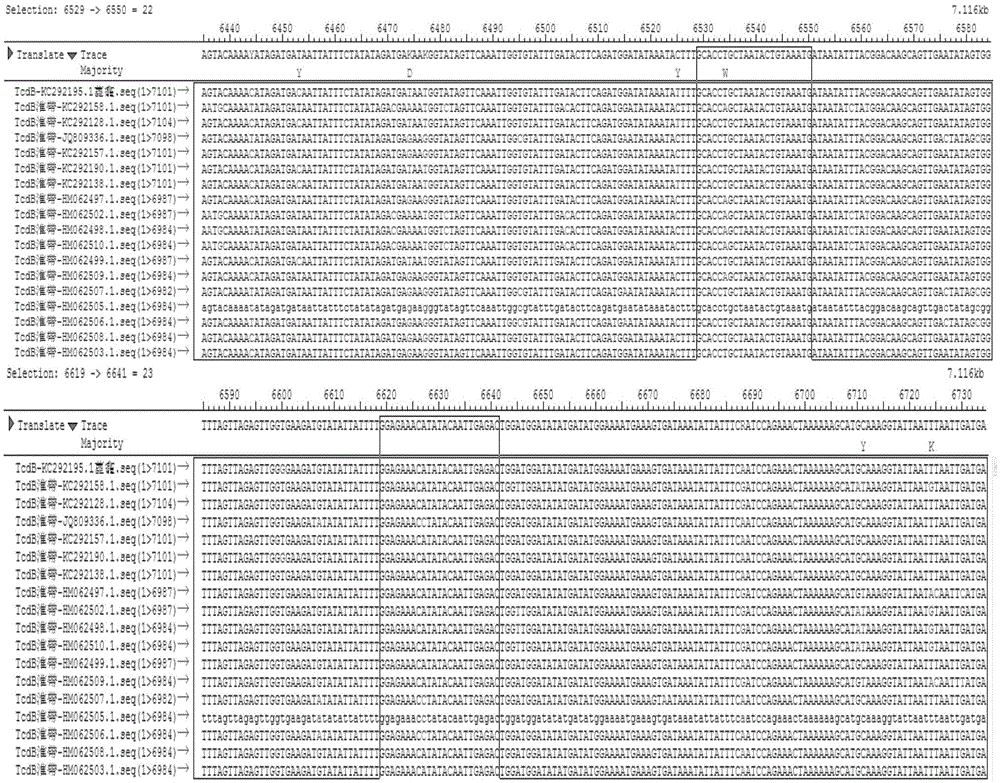
[0070] through the NCBI database and http: / / blast.ncbi.nlm.nih.gov / Blast.cgi Online comparison software to find out the conserved sequences of tcdA gene and tcdB gene among different strains. The sequence of tcdB gene among different strains was very different, and the partial sequence similarity rate was only 94.9%. figure 1 Shown is the full-length sequence conservation analysis of the tcdB gene of different strains, and the boxed position is the position of the outer and inner primer sequence prepared by the primer tcdB quality control product. figure 2 The framed position is the qPCR primer and probe position of tcdB, so the conserved sequence of the tcdB gene was selected with the help of the Saqman software in the DNAStar software package. Finally, the primers were designed with the software PrimerPremier5.0, and the following speci...
Embodiment 3
[0125] Sensitivity comparison between this kit and the existing Clostridium difficile toxin A / B detection kit
[0126] In order to determine the sensitivity of the specific operation method of qPCR described in Example 2, we used the initial concentration of the quality control product as 2.19×10 8 copies / μL(tcdA) and 2.29×10 8 The template concentration of copies / μL (tcdB) was used to prepare the initial concentration of the standard curve, and was serially diluted with EASYDilution Absolute Quantitative Diluent in a 10-fold concentration gradient to detect the minimum detection limit of the qPCR system. In addition, primers from other literatures for detecting C.difficile toxin genes tcdA and tcdB were selected, and the system established by this method was used for qPCR to compare the sensitivity of this primer and the primers in other literatures (TcdCDoesNotSignificantlyRepressToxinExpressioninClostridiumdifficile630DErm).
[0127] Judgment criteria for results: Entero...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com