DNA quantitative method based on rolling circle amplification
A technology of rolling circle amplification and rolling circle amplification reaction, which is applied in the field of DNA quantification, can solve the problems of limited application, decline, and inability to distinguish free nucleotides of double-stranded DNA, and achieves wide concentration range, low error, and high sensitivity. high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
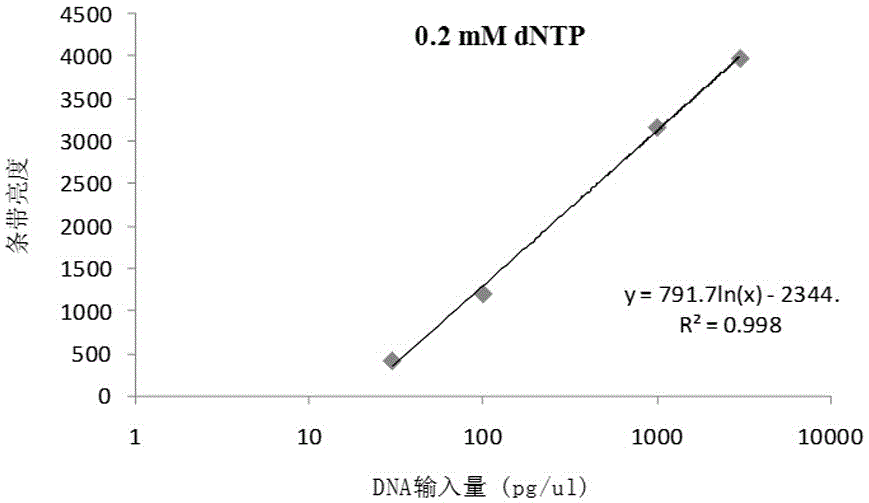
[0032] Example 1 RCA quantification of pGEX4T-1 (4969bp)
[0033] Prepare 150μL sample buffer and 150μL reaction buffer; sample buffer includes random primer, Tris-HCl (400mM, pH8.0), KCl (160mM); reaction buffer includes Tris-HCl (125mM, pH7.5), MgCl 2 (25mM), (NH 4 ) 2 SO 4 (25 mM), DTT (10 mM), dNTPs (used at a final concentration of 0.2 mM).
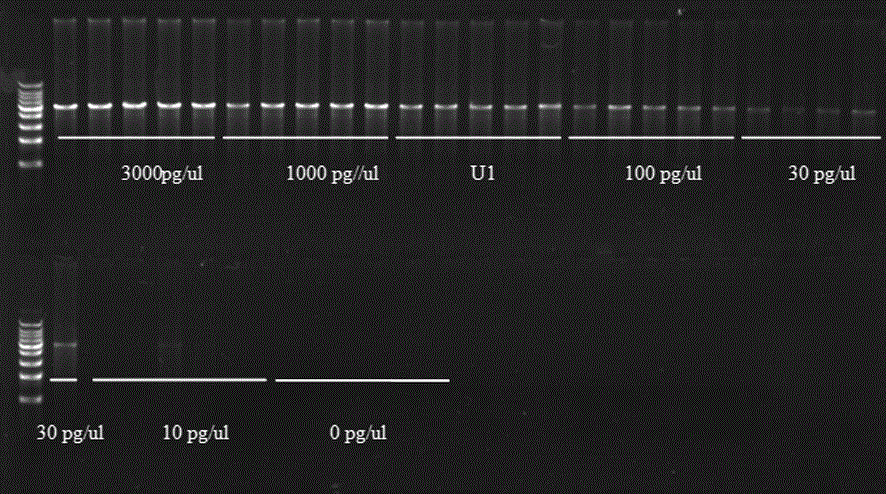
[0034]The sample buffer was added to 35 wells, 4 μL of sample buffer in each well; then pGEX4T- 2 μL of 1’s double-distilled aqueous solution, five groups of each were added to 4 μL of sample buffer, and five groups of 300 pg / μL of pGEX4T-1 double-distilled aqueous solution (called U1) 2 μL were added to 4 μL of sample buffer at the same time. Amplify, react at 95°C for 3 minutes, and centrifuge instantly; then add the reaction buffer to 35 wells, 4 μL of reaction buffer per well, and simultaneously react at 30°C for 3 hours; and finally react at 65°C for 10 minutes to obtain three. Fifteen groups of rolling circle amplification...
Embodiment 2
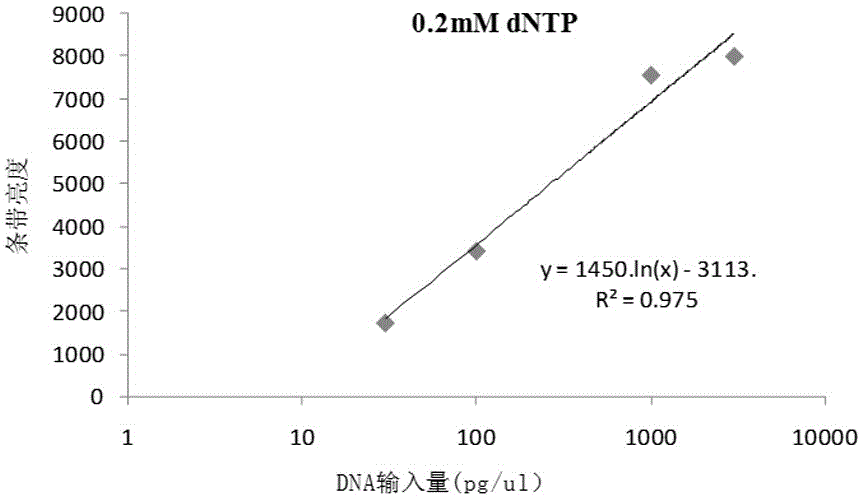
[0038] Example 2 RCA quantification of pUC19 (2686bp)
[0039] Prepare 160μL sample buffer and 160μL reaction buffer; sample buffer includes random primer, Tris-HCl (400mM, pH8.0), KCl (160mM); reaction buffer includes Tris-HCl (125mM, pH7.5), MgCl 2 (25mM), (NH 4 ) 2 SO 4 (25 mM), DTT (10 mM), dNTPs (used at a final concentration of 0.2 mM).
[0040] The sample buffer was added to 32 wells, 4 μL of sample buffer in each well; then the mass concentrations were 0pg / μL, 3pg / μL, 10pg / μL, 30pg / μL, 100pg / μL, 1000pg / μL, 3000pg / μL, respectively. 2 μL of double-distilled aqueous solution of pUC19, four groups were added to 4 μL of sample buffer, and 2 μL of four groups of 320 pg / μL double-distilled aqueous solution of pUC19 (referred to as U2) were added to 4 μL of sample buffer, and rolled at the same time. Loop amplification, react at 95°C for 3 minutes, and centrifuge instantly; then add the reaction buffer to 32 wells, 4 μL of reaction buffer per well, and react at 30°C for 3 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com