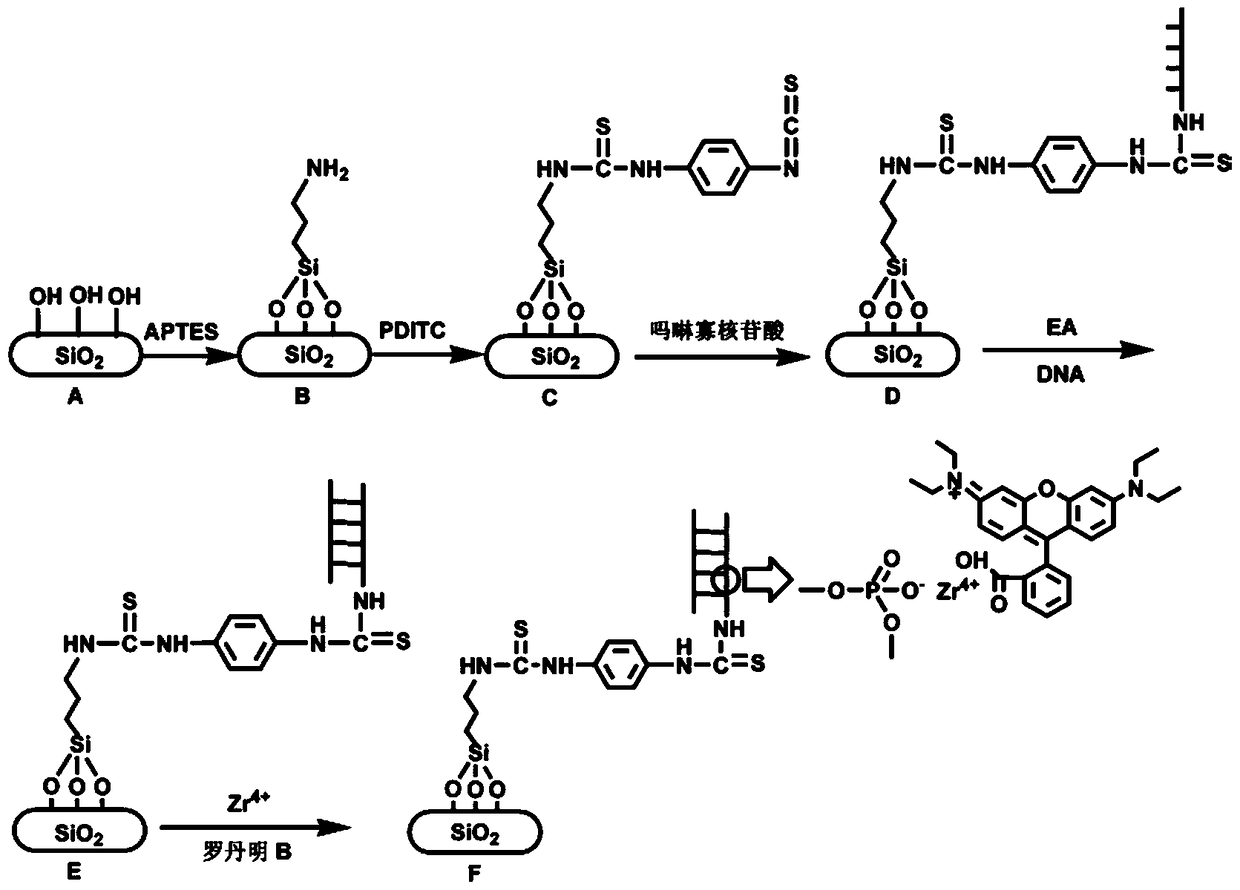
A highly selective DNA fluorescence analysis method based on morpholino oligonucleotide functionalized silicon chip
A morpholino oligonucleotide, a high-selectivity technology, applied in the field of high-selectivity DNA fluorescence analysis, can solve the problems of difficult single nucleotide polymorphism detection, high background noise in electrochemical methods, and low hybridization efficiency. Achieve good linear response, simple method, and short analysis time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
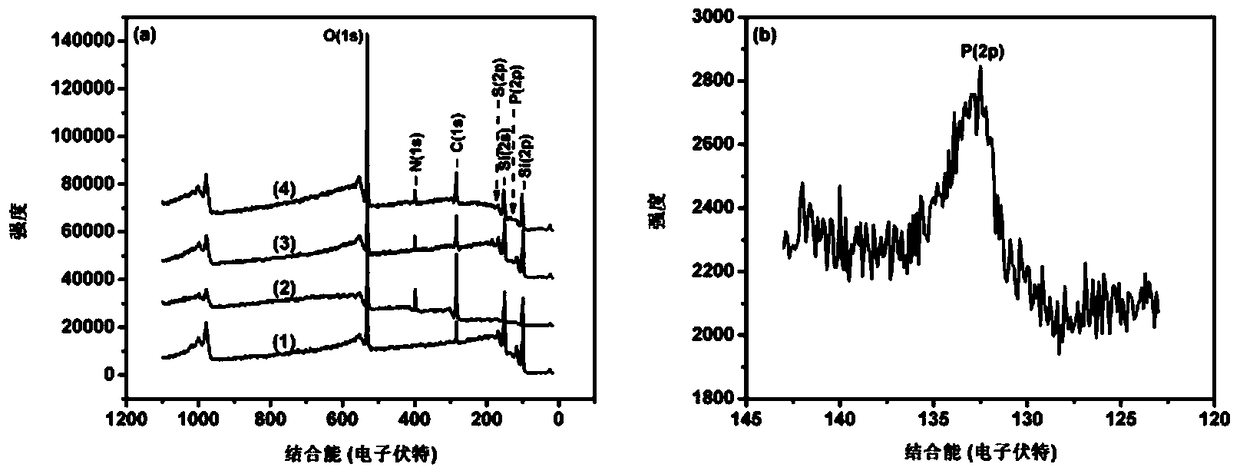
Image
Examples
Embodiment 1
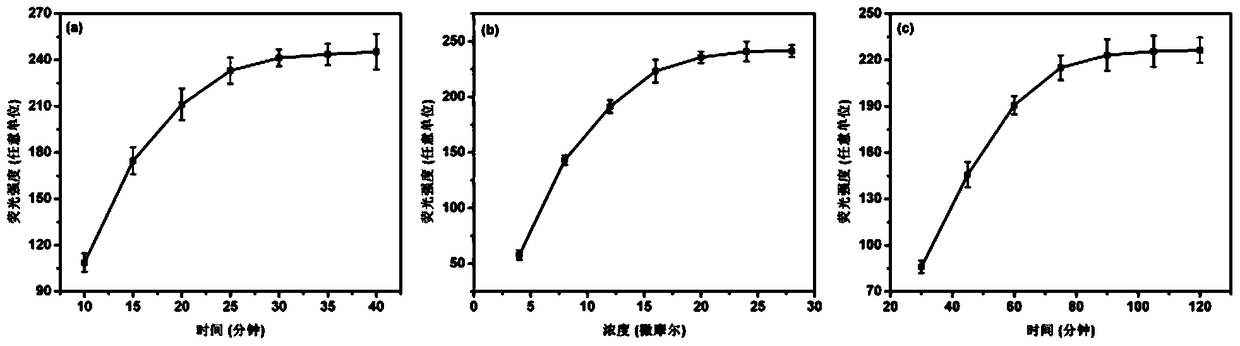
[0046] Example 1: The effect of rhodamine B on the detection results of DNA fluorescence analysis of the present invention under different reaction times.
[0047] Using the highly selective DNA fluorescence analysis based on the morpholine oligonucleotide functionalized silicon-based chip of the present invention, the morpholine oligonucleotide (sequence: 5'-H 2 N-AAC CAT ACA ACC TAC TAC CTC A-3') as capture probe, fully complementary DNA fragment (sequence: 5'-TGA GGT AGT AGG TTG TAT GGTT-3') as target DNA fragment, all operation steps All as described in above-mentioned embodiment 1, wherein, the concentration of morpholine oligonucleotide is 1 μ mol / L, and the concentration of DNA fragment solution to be tested (solvent is 1 * TE buffer) is 0.1nmol / L, hybridization time and rhodan The concentration of rhodamine B remains excessive, and the rhodamine B reaction time is changed to be 10, 15, 20, 25, 30, 35, 40min successively, and the influence of different rhodamine B react...
Embodiment 2
[0048] Example 2: Effects of different rhodamine B concentrations on the detection results of the DNA fluorescence analysis of the present invention.
[0049] Using the highly selective DNA fluorescence analysis based on the morpholine oligonucleotide functionalized silicon-based chip of the present invention, the morpholine oligonucleotide (sequence: 5'-H 2 N-AAC CAT ACA ACC TAC TAC CTC A-3') as capture probe, fully complementary DNA fragment (sequence: 5'-TGA GGT AGT AGG TTG TAT GGTT-3') as target DNA fragment, all operation steps All as described in above-mentioned embodiment 1, wherein, the concentration of morpholine oligonucleotide is 1 μ mol / L, the solution (solvent is 1×TE buffer) concentration of DNA fragment to be tested is 0.1 nmol / L, and hybridization time keeps excessive , change the Rhodamine B concentration to 4, 8, 12, 16, 20, 24, 28 μmol / L in turn, and analyze the influence of different Rhodamine B concentrations on the nucleic acid fluorescence analysis detec...
Embodiment 3
[0050] Example 3: Effects of different hybridization reaction times on the detection results of the DNA fluorescence analysis of the present invention.
[0051] Using the highly selective DNA fluorescence analysis based on the morpholine oligonucleotide functionalized silicon-based chip of the present invention, the morpholine oligonucleotide (sequence: 5'-H 2 N-AAC CAT ACA ACC TAC TAC CTC A-3') as capture probe, fully complementary DNA fragment (sequence: 5'-TGA GGT AGT AGG TTG TAT GGTT-3') as target DNA fragment, all operation steps All as described in above-mentioned embodiment 1, wherein, the concentration of morpholine oligonucleotide is 1 μ mol / L, the solution (solvent is 1×TE buffer) concentration of DNA fragment to be tested is 0.1nmol / L, changes DNA to be tested The hybridization reaction time of the fragment and the capture probe morpholino oligonucleotide is 30, 45, 60, 75, 90, 105, and 120 min in sequence, and the influence of different hybridization reaction times...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com