Saccharomyces cerevisiae gene engineering bacteria for producing succinic acid and application thereof
A technology of genetically engineered bacteria and Saccharomyces cerevisiae, applied in the field of Saccharomyces cerevisiae genetically engineered bacteria, can solve the problems of difficulty in accumulation and loss of carbon flow, and achieve the effect of achieving accumulation and reducing the loss of carbon flow
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
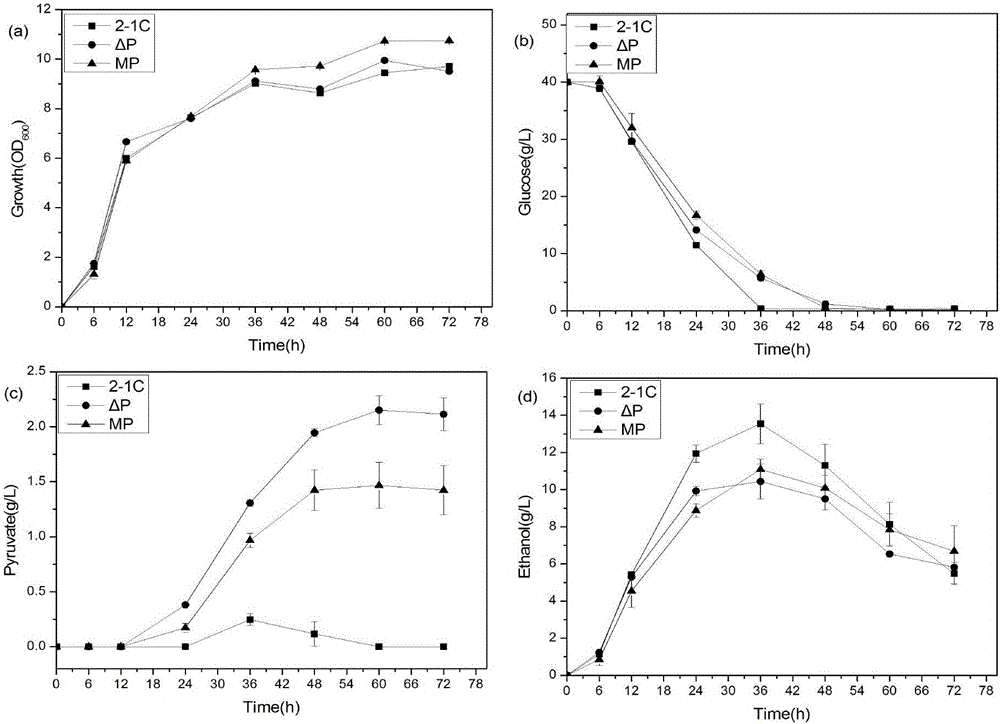
[0028] Example 1 Construction of low-yield ethanol and accumulation of succinic acid Saccharomyces cerevisiae engineering bacteria
[0029] On the basis of the wild type, the pyruvate decarboxylase gene PDC1 was replaced with the malate dehydrogenase gene RoMDH from Rhizopus oryzae. On this basis, the key enzyme gene SDH2 in the succinate metabolic pathway was knocked out.
[0030] 1. Strains and plasmids
[0031] Saccharomyces cerevisiae (S.cerevisiae) CEN.PK2-1C was purchased from EUROSCARF (http: / / web.uni-frankfurt.de / fb15 / mikro / euroscarf / data / cen.html), and its genotype was MATa; ura3- 52; trp1-289; leu2-3,112; his3Δ1; MAL2-8 C;SUC2. For the construction method of knockout cassette template pUG27 plasmid (containing HIS3 marker gene) and Cre expression plasmid pSH47 (used for marker recovery), please refer to the literature "Guldener U, Heck S, Fielder T, et al. A new efficient gene disruption cassette for repeated use in budding yeast[J].Nucleic Acids Res,1996,24(13):...
Embodiment 2
[0038] Embodiment 2 fermentation method produces succinic acid
[0039] The seeds of genetically engineered bacteria cultured at 30°C and 220rpm for 24 hours were used to start the OD 600 The inoculum size of =0.2 was transferred to the fermentation culture based on the conditions of 30°C and 220rpm for 96h, and samples were taken at regular intervals to measure OD, and 1mL samples were taken at the same time and centrifuged for storage to measure the production of ethanol and succinic acid.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com