Application of soybean MYB transcription factor gene in improvement of soybean isoflavone biosynthesis
A technology of soybean isoflavones and transcription factors, applied in the field of genetic engineering, can solve problems such as inability to obtain results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1, the cloning of soybean GmMYB9 gene
[0026] RNAiso Plus (purchased from TaKaRa) was used to extract total RNA from 20-day embryos of soybean variety Williams 82, and the integrity of RNA was detected by 1% agarose electrophoresis. cDNA was synthesized according to Reverse Transcriptase M-MLV (RNase H - ) instructions. Primers were designed according to the full-length sequence of GmMYB9, and the primers were:
[0027] GmMYB9-F:AACAACATAGAGAGCCATAATACCC
[0028] GmMYB9-R: CATAGACTCCTCTTTCAAAACACCTC
[0029] Perform PCR amplification according to the reaction system in Table 1:
[0030] Table 1 PCR amplification reaction system
[0031]
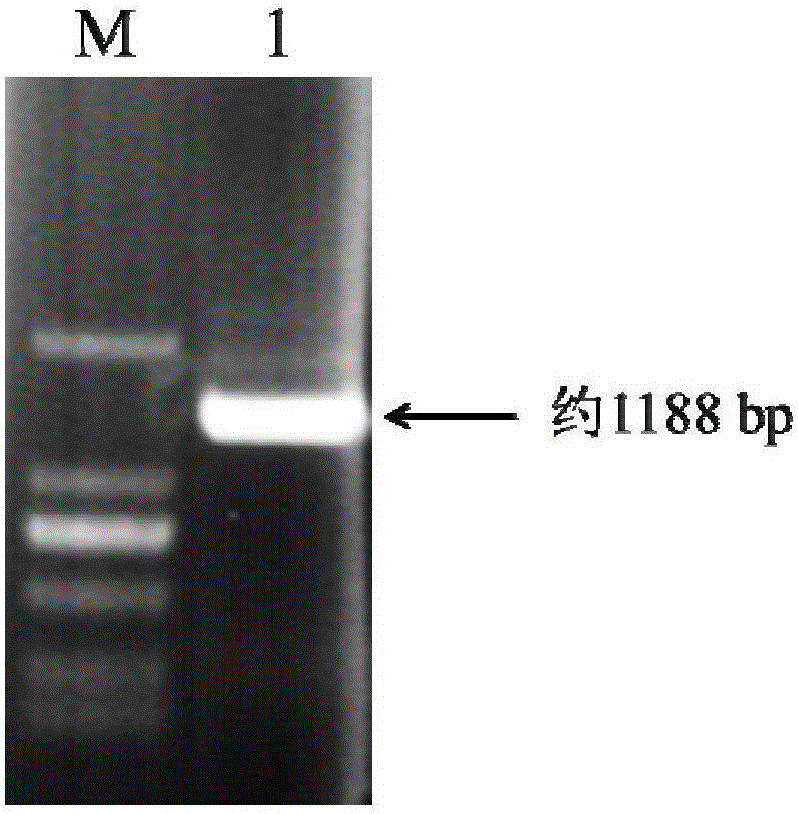
[0032] The PCR reaction program was: 95°C for 4min; 30 cycles of 94°C for 30s, 57°C for 50s, and 72°C for 90s; 72°C for 10min. The GmMYB9 gene was obtained by PCR amplification, such as figure 1 As shown, it consists of 1188 base pairs, and the reading frame is from the 1st to the 1188th base at the 5' end, encodi...
Embodiment 2
[0033] Embodiment 2, expression of soybean GmMYB9 gene in yeast
[0034] Primers containing EcoR I and Sal I restriction sites were designed at the 5' end and 3' end of the gene, respectively, and the plasmid pMD18-T-GmMYB9 identified correctly by sequencing was used as a template for PCR amplification. pMD18-T was purchased from TaKaRa Corporation. Primers are as follows (restriction sites are underlined):
[0035]GmMYB9-HF: 5'- CCGGAATTC ATGGGAAGGAGTCCTTGC-3', EcoR I
[0036] GmMYB9-HR: 5'- ACGCGTCGAC CTACAGATACTGAATGTA-3', Sal I
[0037] After the amplified product was purified by an agarose gel DNA purification and recovery kit (purchased from BioTeke Company), it was digested with EcoRI and Sal I, and after recovery and purification, it was mixed with pGBKT7 ( purchased from clontech) carrier ligation. The ligated product was transformed into Escherichia coli (E.coli) DH5α, and after verification, it was transformed into yeast AH109 (purchased from clontech) for e...
Embodiment 3
[0040] Example 3. Research on subcellular localization of soybean GmMYB9 gene
[0041] The stop codon of GmMYB9 was removed, and the fusion gene of GmMYB9 and green fluorescent protein gene eGFP was constructed by overlapping extension PCR (SOE-PCR), and Xba I and SacI restriction sites were designed at the 5' and 3' ends respectively. Primers, using the correctly identified pMD18-T-GmMYB9 and peGFP-X1 as templates, and using O-F3 and O-R3, O-F4 and O-R4 as paired primers for common PCR amplification, PCR products were recovered and made Good mark. Primers are as follows (restriction sites are underlined):
[0042] GmMYB9-SOE-F(O-F3):5'- GCTCTAGA ATGGGAAGGAGTCCTTGC-3',Xba I
[0043] GmMYB9-SOE-R(O-R3):
[0044] 5'-CACCAT CAGATACTGAATGTACTT-3' (the wavy line represents
[0045] Linker between target gene and reporter gene)
[0046] eGFP-SOE-F(O-F4):
[0047] 5'-GTATCTG ATGGTGAGCAAGGGCGAG-3'
[0048] eGFP-SOE-R(O-R4):5'- CGAGCTC TTACTTGTACAGCTCGTC-3', SacI
[004...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com