Antibody discovery method based on hybridoma technique and high-throughput sequencing
A hybridoma and high-throughput technology, applied in the field of biomedicine, can solve the problems of difficulty in obtaining high-affinity monoclonal antibodies, increase the possibility of obtaining functional antibodies, and the small range of antibody cell line screening, so as to overcome the blindness of pairing, Increased likelihood, highly reproducible effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
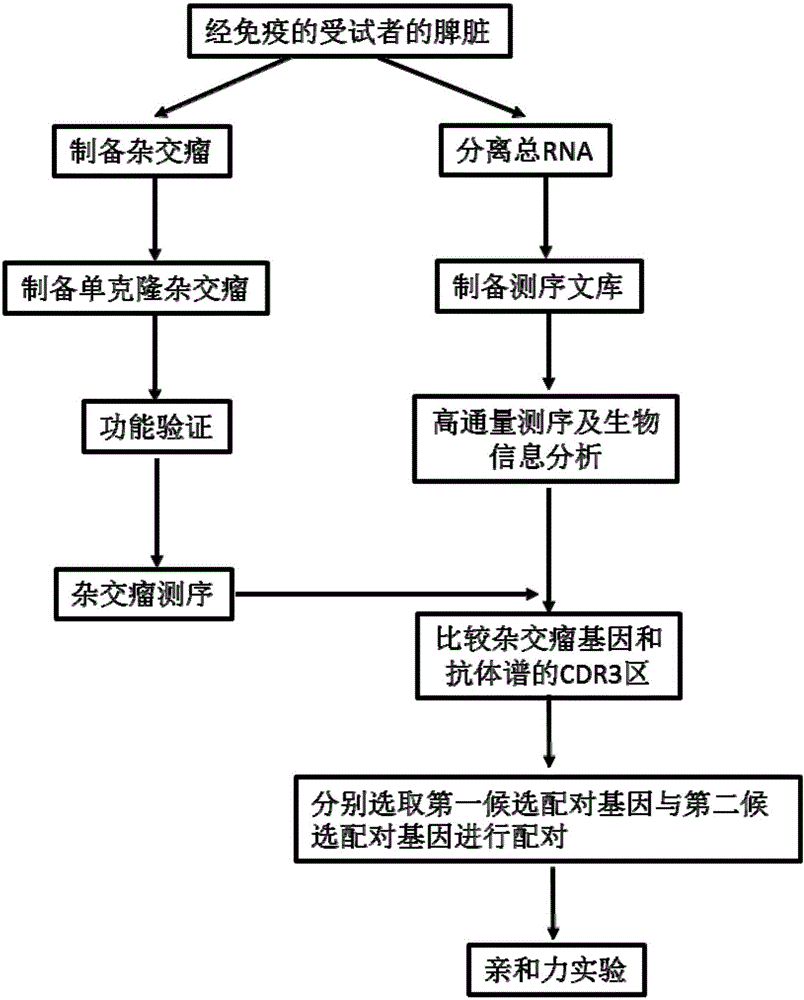
[0060] Example 1 Isolation and Identification of Hybridoma Antibody Sequences
[0061] Use the following procedure:
[0062] (1) Expand and culture myeloma cells so that they are in the logarithmic growth phase when confluent. On the day of fusion, cells were collected, counted, and set aside.
[0063] (2) Take the hemagglutinin HA antigen polypeptide (hereinafter represented by the code GW01) and the immune checkpoint PD-L1 antigen polypeptide (hereinafter represented by the code GW-02) whose serum titer reaches the standard, and prepare hybridization with two immunized mice each Tumors were named GW01-1, GW01-3 and GW02-3, GW02-5, respectively. Eyeballs of the mice were removed to collect blood, and the serum was separated as a positive control serum for antibody detection. The mice were killed by pulling the neck, and the spleens of the mice were taken aseptically, the splenocytes were separated and the red blood cells were lysed to obtain pure B lymphocytes. Cell fusio...
Embodiment 2
[0066] Example 2 Preparation of antibody sequencing library and analysis of antibody sequences
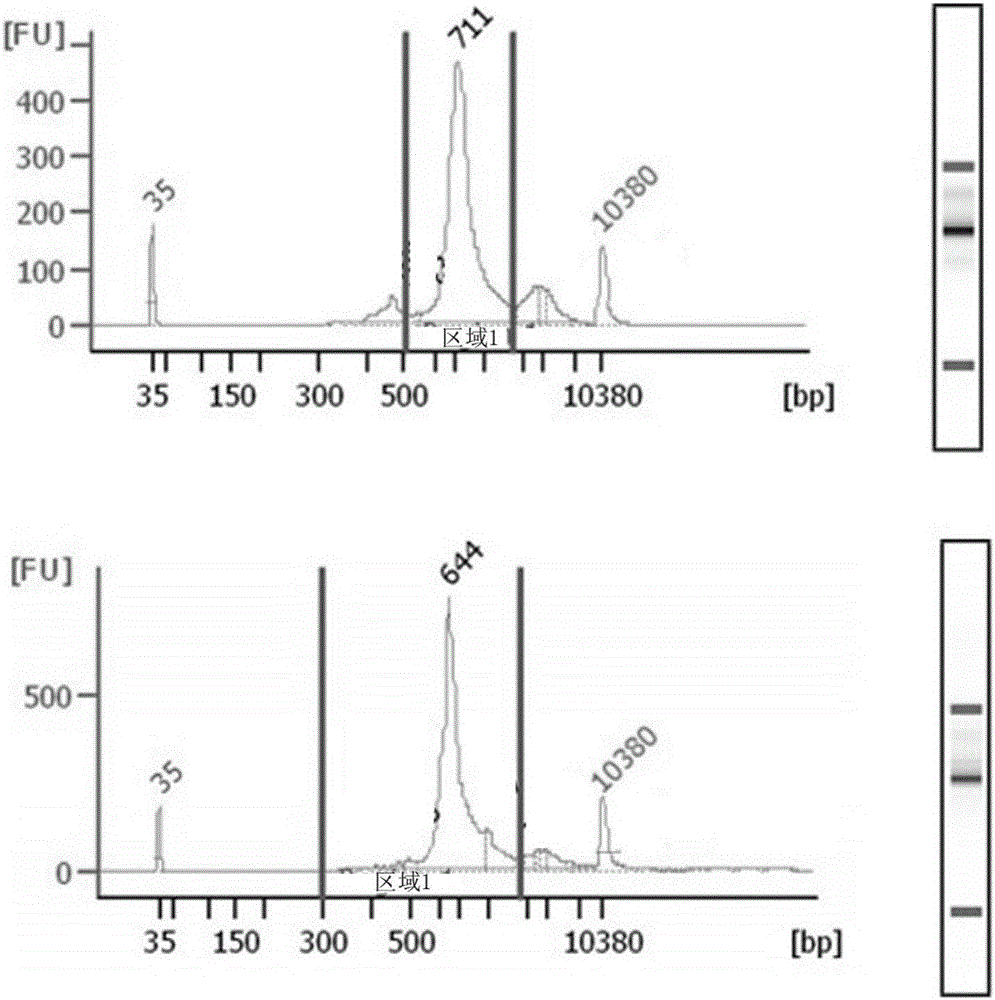
[0067] The remaining spleen tissue of the hybridoma prepared in Example 1 and the bone marrow cells from the same immunized mouse were used to prepare the antibody sequencing library.
[0068] Specifically, the spleen tissue was ground in liquid nitrogen to extract total RNA, and the specific extraction procedure was carried out according to the manufacturer's instructions of the extraction reagent; the tibia and femur were collected from the same immunized mouse to remove muscle and fat tissue, and the Inject Trizol into the bone to wash out the bone marrow and collect it in a centrifuge tube to extract total RNA. The specific extraction procedure is carried out according to the manufacturer's instructions of the extraction reagent.
[0069] Using antibody variable region upstream primers and heavy chain and light chain variable region downstream primers, using the sample 5' RACE ...
Embodiment 3
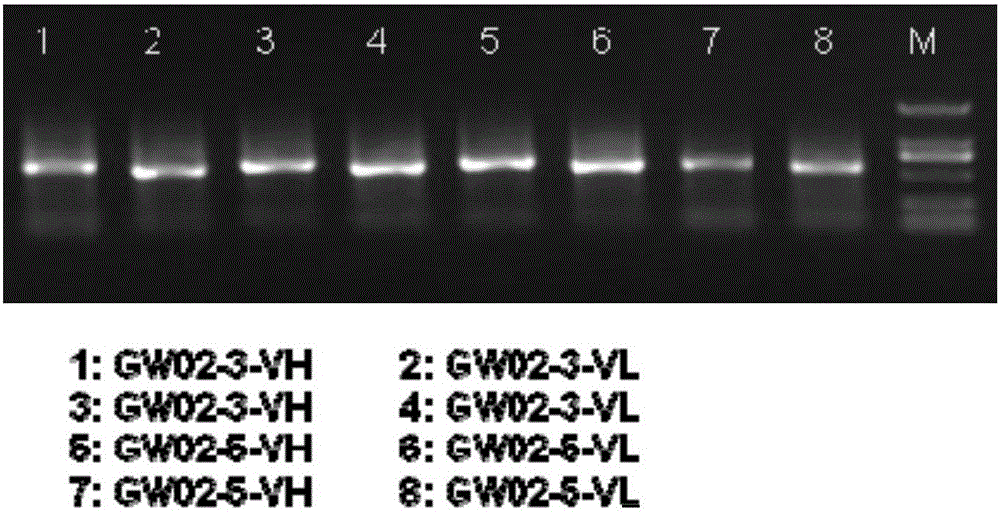
[0092] Example 3 Comparison of Hybridoma and Antibody Variable Region Gene Profile Sequences and Selection of Candidate Matching Sequences
[0093] A total of 18 GW02 and 23 GW01 monoclonal hybridoma cell lines were analyzed, and the variable region of the hybridoma antibody gene was submitted for IgBlast to determine the information of CDR3H and CDR3L fragments. Comparing CDR3H and CDR3L with the corresponding antibody variable region gene profiles, most of the hybridoma CDR3H and CDR3L are included in the antibody variable region gene profiles, and the number of sequencing entries corresponding to some CDR3s exceeds 1% of the total number of sequencing entries. Belongs to high abundance CDR3. The number of high-abundance CDR3 and the proportion of hybridoma CDR3 in the antibody variable region gene profile are shown in Table 9 below.
[0094] Table 9
[0095]
[0096] In Table 9, CDR3>1%: the number of hybridoma CDR3s exceeding 1% of the number of antibody sequencing li...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com