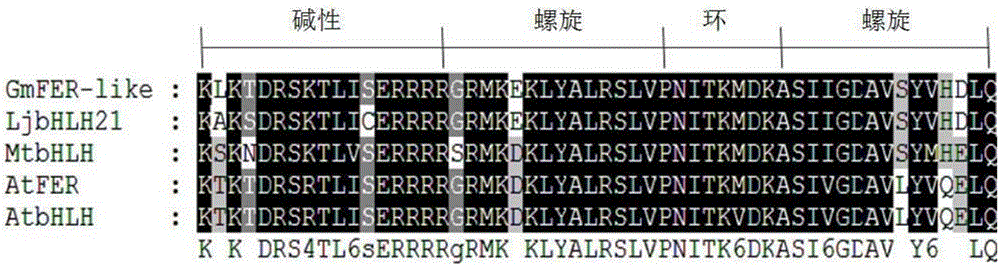
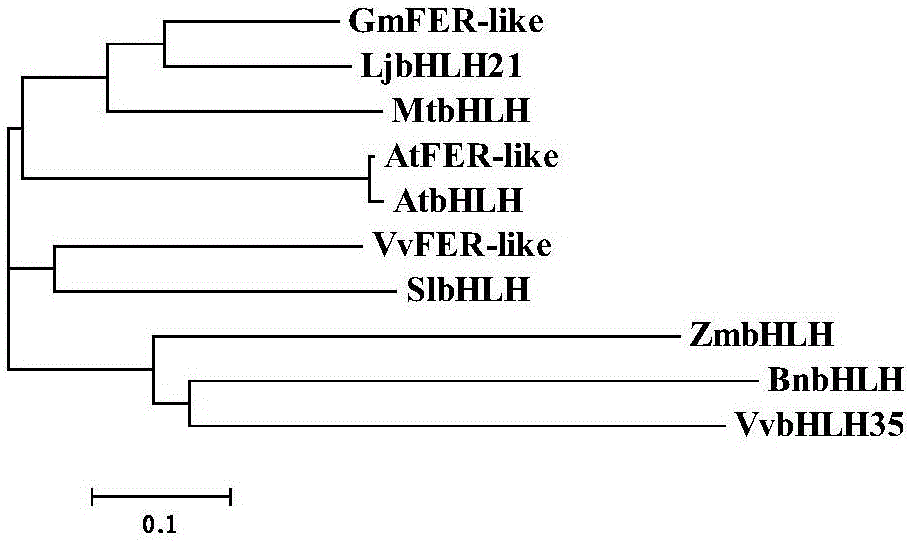
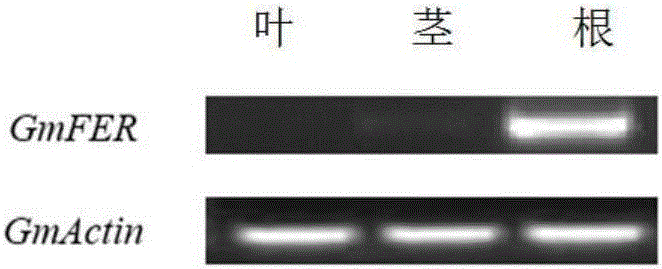
Soybean bHLH transcription factor gene GmFER and encoded protein and application thereof
A technology for transcription factors and transgenic plants, which is applied in the application field of soybean bHLH transcription factor gene GmFER, tolerance, and can solve the problems of reducing transport, and no report discloses the related application of soybean bHLH transcription factor gene, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Cloning of soybean bHLH transcription factor gene GmFER.
[0026] Utilize the method that bioinformatics and routine polymerase chain reaction (PCR) combine to clone the dna sequence of soybean GmFER gene, specific method is as follows:
[0027] The root system of soybean variety Williams 82 was used as experimental material, and total RNA was extracted using SV Total RNAlysis Kit (Promega, USA). According to the method provided by TaKaRa Company, using PrimeScript TM Reverse transcriptase synthesizes cDNA, and uses it as template to carry out PCR amplification, a pair of primers P1 (as shown in SEQ ID NO:3, synthesized by Shanghai Bioengineering Company) and P2 (as shown in SEQ ID NO:3) and P2 (as shown in SEQ ID NO:3) used in the PCR amplification process NO:4, synthesized by Shanghai Bioengineering Company) was designed and synthesized according to the known GmFER sequence in NCBI:
[0028] P1: 5'-ATGGATGTTCACGAAGACACACTCA-3';
[0029] P2: 5'-TCAAGCAGGA...
Embodiment 2
[0031] Example 2: Spatiotemporal distribution of GmFER transcripts and the effect of Cd stress on GmFER gene expression.
[0032] Use the method of semi-quantitative PCR to detect the tissue distribution of GmFER in soybean plants, and take the following measures to ensure the reliability of the detection results: Use the amplification grade DNase I to digest and extract a small amount of genomic DNA contamination in the total RNA , in order to detect whether the genomic DNA is eliminated, a part of the total RNA sample digested with DNase I can be taken out for conventional PCR reaction. When it was found that no amplified band was generated, the reverse transcription step was performed again, and at the same time, in order to further verify whether the band amplified by real-time quantitative PCR was GmFER, the PCR product was gel-cut and recovered for sequencing. The specific method is as follows: total RNA is extracted from materials such as soybean roots, stems, leaves, f...
Embodiment 3
[0038] Example 3: Construction of yeast expression vector containing GmFER gene and detection of tolerance of transgenic yeast to Cd.
[0039] (1) Construction of yeast expression vector:
[0040] Using the full-length sequence of GmFERcDNA as a template, forward primer P5 (shown in SEQ ID NO: 7, synthesized by Shanghai Bioengineering Co.) and reverse primer P6 (shown in SEQ ID NO: 8) containing restriction sites , synthesized by Shanghai Bioengineering Company), PCR amplified the full-length sequence of GmFERcDNA with BamHI recognition site added at the 5' end and SmaI recognition site added at the 3' end. The nucleotide sequences of the primers were as follows:
[0041] P5: 5′-CGC GGATCC ATGGATGTTCACGAAGACACACTCA-3' (the underlined base is the recognition site of the restriction endonuclease BamHI);
[0042] P6: 5′-TCC CCCGGG TCAAGCAGGAAAAGATGCCACGAAT-3' (bases underlined are recognition sites for restriction endonuclease SmaI).
[0043] After the reaction, the PCR amp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com