SUMO and SUMO protease encoding gene and application thereof
A protease and gene technology, applied in the fields of genetic engineering and protein engineering, to achieve high expression, increase expression, and broaden application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0045] A kind of preparation method of the recombinant vector of pET28b-sumo (S) and pMF-ulp (S) of the present invention, comprises the steps:
[0046] (1) Artificially synthesizing the genes shown in SEQ ID NO: 1 and SEQ ID NO: 3 in the sequence table respectively;
[0047] (2) Construct the recombinant vectors of pET28b-sumo (S) and pMF-ulp (S) respectively; the pMF-ulp (S) recombinant vector is selective to the vector, and the present invention proves that ulp (S) is constructed in pET28b Most of the expression in the vector is inclusion body precipitation, while soluble expression can be achieved in the pMF vector.
[0048] The expression vector of the Saccharomyces cerevisiae gene sumo (S) is pET28b, the expression vector of the SUMO protease gene ulp (S) is a pMF vector, and the host bacterium is Escherichia coli DH5α;
[0049] Using DNA recombination technology, insert the sumo(S) gene shown in the sequence table SEQ ID NO: 1 into the Nde Ⅰ and BamH Ⅰ enzyme digestion...
Embodiment 1
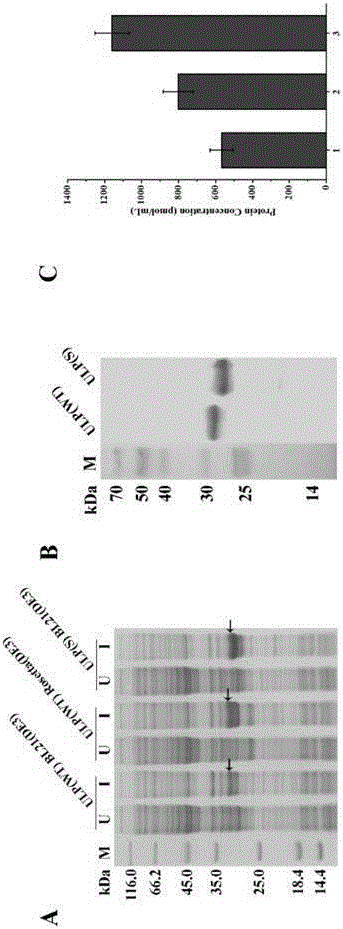
[0064] Construction, expression, detection and expression measurement of sumo(S) and ulp(S) recombinant vectors
[0065] The sumo(S) gene and pET28b vector were digested with Nde Ⅰ and BamH Ⅰ, and the ulp(S) gene and pMF vector were digested with BamHI and HindⅢ. After gel recovery, the corresponding gene and vector were ligated with T4 DNA ligase at 16°C for 5 hours. , the ligation product was transferred into E. coli DH5α competent cells, after incubation and refolding, smear a resistant plate and culture at 37°C for 12 hours, then pick a single colony, and culture in 5mL Luria-Bertani medium at 37°C, 220rpm for 12 hours Plasmids were extracted and sequenced after enzyme digestion and verification.
[0066] The correctly sequenced pET28b-sumo(S) and pMF-ulp(S) plasmids were transferred into BL21(DE3) competent cells, the control plasmids were pET28b-sumo(wt) and pET28b-ulp(wt), and the control plasmids Transform into BL21(DE3) and Rosetta(DE3) competent cells, pick a single...
Embodiment 2
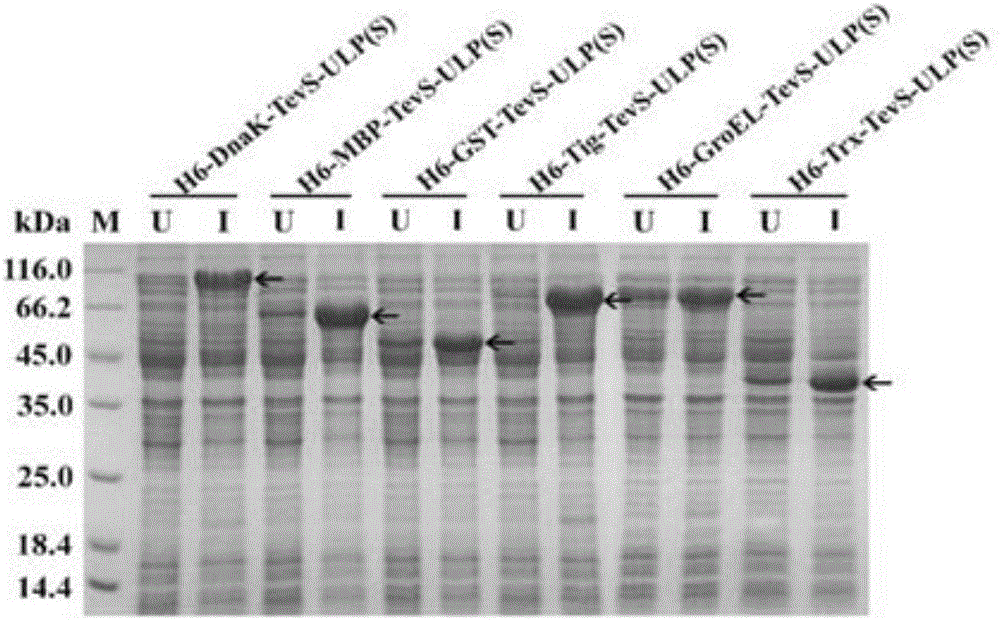
[0073] Construction and expression of ulp(S) recombinant vector containing six kinds of fusion tags
[0074] The six tags of dnaK, mbp, gst, tig, groEL, trx and pET28b-tevS were digested with Nde Ⅰ and Bgl Ⅱ and then ligated and transformed into DH5α competent cells. The vector and ulp(S) were digested with BamH Ⅰ and Hind Ⅲ and then ligated. After the transformation and digestion were verified, the six plasmids were transferred into BL21(DE3) competent cells. -Bertani medium 37 ℃, 220rpm shaking culture, bacteria grow to OD 600 At 0.6, add IPTG with a final concentration of 0.5mmol / L and induce at 28°C for 12h, then collect the bacteria and sonicate them, freeze and centrifuge the supernatant protein samples for SDS-PAGE electrophoresis. Such as image 3 shown. The results showed that the expression of ULP(S) with tag fusion was good.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com