Lead ion visual detection method
A detection method and technology for lead ions are applied in the field of rapid visual detection of lead ions, which can solve the problems of interfering lead ion detection and low detection sensitivity, and achieve the effects of high sensitivity and rapid detection, high detection sensitivity, and improved use efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0046] 1) Preparation of gold-labeled probe:
[0047] Preparation of the first gold-labeled probe: Add the sulfhydryl-modified first single-stranded DNA to the nano-gold solution, react for 24 hours, then add phosphate buffer and sodium dodecylsulfonate, mix at room temperature for 30 minutes, add chloride Aging at room temperature with sodium for 2 days, centrifuging at 4°C for 25 min, discarding the supernatant, and resuspending the precipitate with resuspension buffer to obtain the prepared first gold-labeled probe;
[0048] Preparation of the second gold-labeled probe: Add the sulfhydryl-modified second single-stranded DNA to the nano-gold solution, react for 24 hours, then add phosphate buffer and sodium dodecylsulfonate, mix at room temperature for 30 minutes, add chloride Aging with sodium at room temperature for 2 days, centrifuging at 4°C for 25 min, discarding the supernatant, and resuspending the precipitate with resuspension buffer to obtain the prepared second gol...
Embodiment 1
[0067] A rapid visual detection method for lead ions, comprising the steps of:
[0068] (1) Preparation of nano gold solution:
[0069] 100mL HAuCl 4 The aqueous solution (0.01%) was poured into a round bottom flask equipped with a reflux condenser and heated to boiling, then 2 mL of trisodium citrate solution (1%) was added to the flask. Continue to heat and reflux under strong magnetic stirring. After the color of the solution gradually changes from colorless and black to deep red, continue heating for 10 minutes, stop heating, cool to room temperature, filter with a 0.22 μm nylon membrane to remove large particles, and store it at temperature Set aside in the refrigerator at 4 °C.
[0070] (2) Preparation of gold standard probe:
[0071] Add 45 μL of thiol-modified first single-stranded DNA and second single-stranded DNA (100 μM, 1OD) to 14 mL of nano-gold solution, react for 24 h, add 100 mmol / L PB (phosphate buffer) and 10% SDS ( sodium dodecyl sulfate) to make the fi...
Embodiment 2
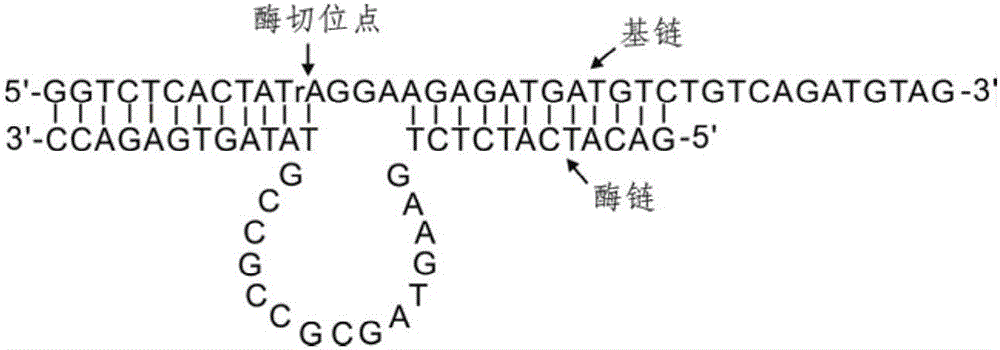
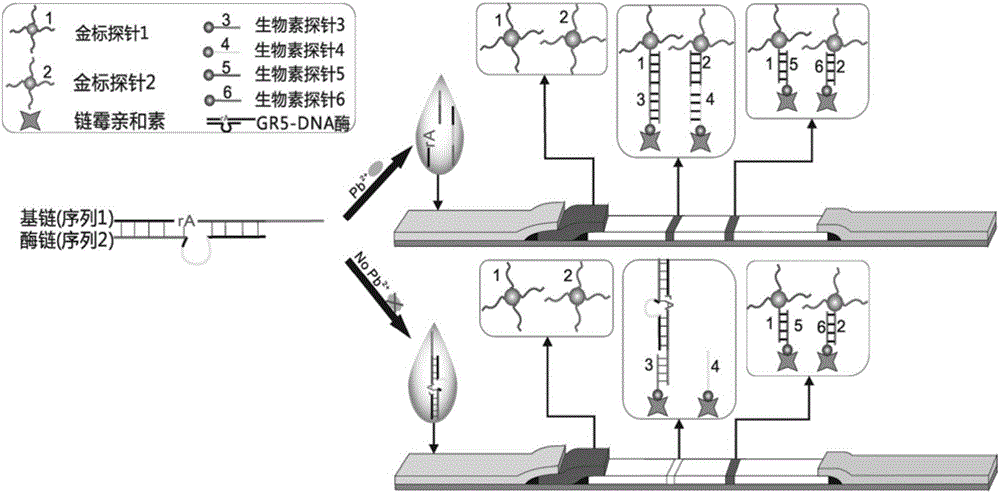
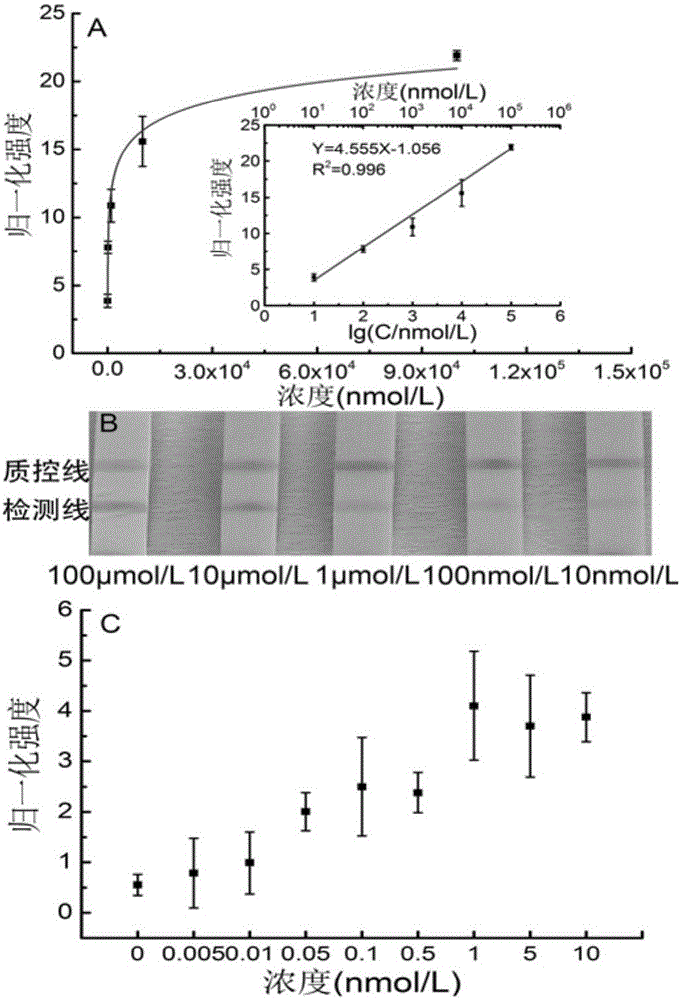
[0085] A kind of lead ion gold label chromatography test strip provided by the invention, such as figure 2 As shown, including the sample pad, gold standard pad, nitrocellulose membrane and absorbent paper carried on the PVC bottom plate in sequence, the gold standard pad is fixed with the first gold standard probe ( figure 2 The gold-labeled probe 1) and the second gold-labeled probe ( figure 2 The gold standard probe 2) in the medium; There are detection line and quality control line on the nitrocellulose membrane, and the third biotin probe is immobilized on the detection line ( figure 2 The biotin probe 3) and the fourth biotin probe ( figure 2 The biotin probe 4) in the quality control line is immobilized with the fifth biotin probe ( figure 2 The biotin probe 5) and the sixth biotin probe ( figure 2 Biotin probes in 6). The base chain GR-5S and the enzyme chain GR-5E are complementary paired, the excess free single chain is adsorbed by graphene oxide, and lead...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com