Benzoyl hydrazine allosteric inhibitor for D-3-phosphoglycerate dehydrogenase and applications of benzoyl hydrazine allosteric inhibitor
A technique of use and medicinal salt, applied in the field of allosteric inhibitors of benzohydrazide D-3-phosphoglycerate dehydrogenase and its use, can solve the problems of no drug effect, no PHGDH inhibitor, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1, the discovery of PHGDH allosteric inhibitor
[0037] 1. Prediction of PHGDH allosteric sites
[0038] The allosteric sites of PHGDH (PDB code: 2G76) were predicted using the protein surface exploration program CAVITY. First, the program probes the surface of the protein by erasing the ball method to find potential binding sites on the protein surface; then, the program uses the empirical formula (CavityScore=(Volume-AdjustVolume) / (SurfaceArea-AdjustSurfaceArea)) to analyze the protein-binding small molecule ability to score. AdjustVolume and AdjustSurfaceArea are related to the hydrophobic area of residues and the number of hydrogen bond acceptors and donors in the predicted sites. by the maximum pK for known binding site-ligand binding pairs D Score and compare with known experimental pK D A good linear correlation was obtained by fitting the values. Therefore, the program can score the predicted binding site-ligand maximum pK according to the abov...
Embodiment 2
[0041] Embodiment 2, the synthesis of allosteric molecule
[0042] 1. Design of PKUMDL-WL-2101 analogs
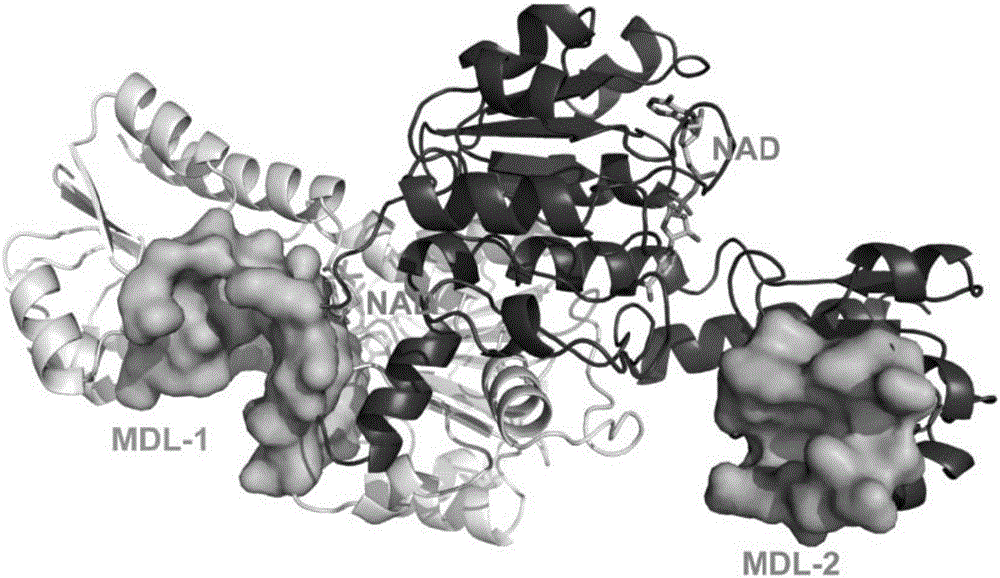
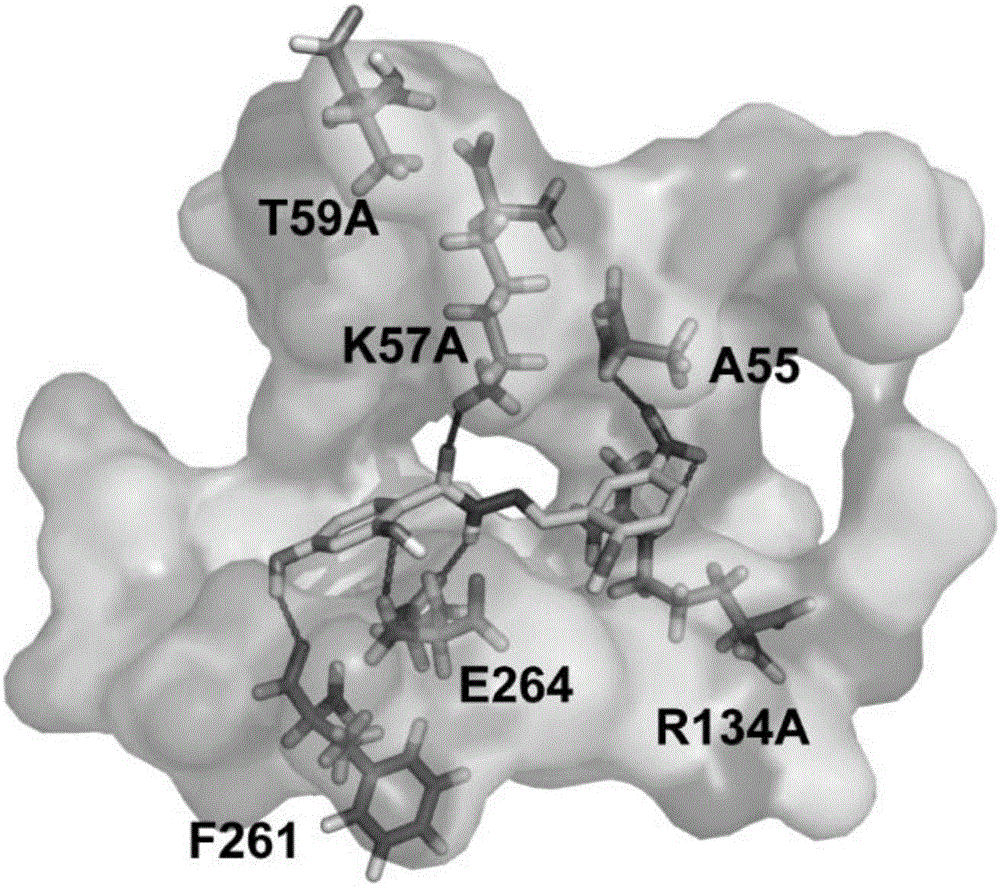
[0043] According to the docking model of the active compound compound (E)-2,4-dihydroxy-N'-(2-hydroxy-5-nitrobenzylidene)benzohydrazide (PKUMDL-WL-2101) bound to PHGDH ( See figure 2 ), the interaction mode between the small molecule and PHDGH can be seen: two benzene rings occupy the hydrophobic cavity in the pocket, and on the benzene ring of the acyl group, the hydroxyl group substituted at position 4 can form a hydrogen bond with phenylalanine at position 261; The substituted hydroxyl can form a hydrogen bond with 264 glutamic acid; the carbonyl oxygen in the hydrazide chain can form a hydrogen bond with 57 lysine, and the hydrazide nitrogen can form a hydrogen bond with 264 glutamic acid; on another benzene ring, The nitro group at position 3 can form hydrogen bonds with arginine at position 134 or alanine at position 55; the ring also has electrostatic interactions...
Embodiment 3
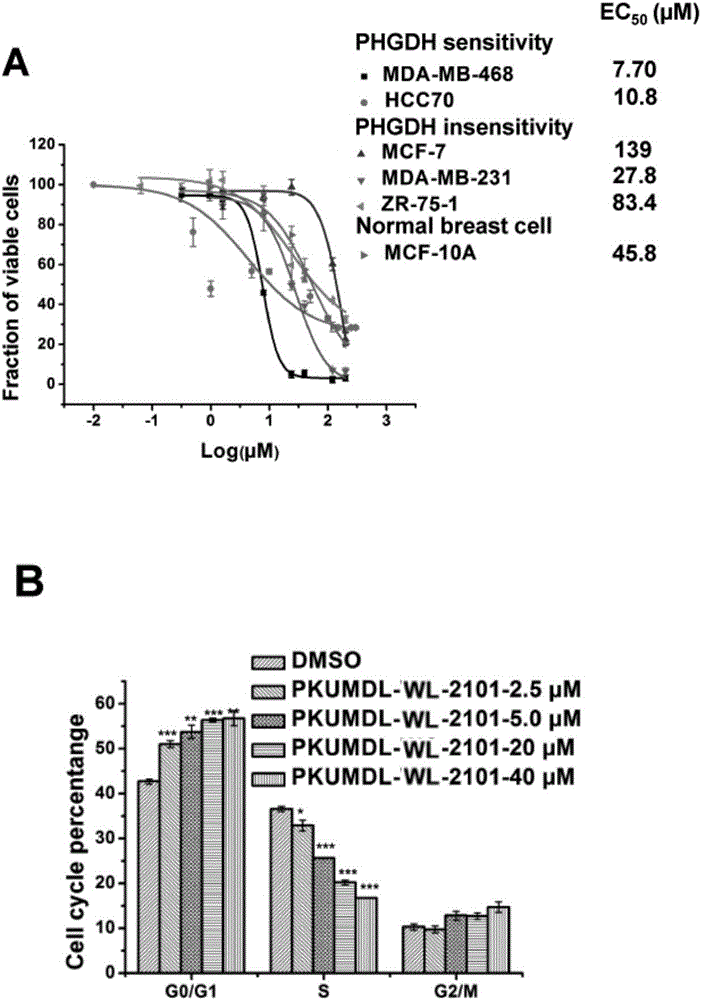
[0094] Example 3, Fluorescence Kinetic Method Determination of PHGDH Enzyme Activity in Vitro of PKUMDL-WL-2101 and Its Analogs
[0095] The determination of PHGDH enzyme activity is realized by detecting the fluorescence emission spectrum of NADH at 456nm. First, PHGDH (final concentration 30 ng / μL) was incubated with HEPES buffer (25 mM, pH 7.1, 400 mM KCl), 5 μM PLP, 0.5 mM αKG, 150 μM NADH and PSAT1 (final concentration 30 ng / μL) in a 96-well plate for 10 minutes. Subsequently, 10 μL of DMSO (control group) or DMSO solution containing small molecules was added, and shaken and balanced at 25° C. at 550 rpm for 5 minutes. The final concentration (v / v) of DMSO was kept at 5% in the enzyme activity test system. Finally, Pser aqueous solution (final concentration 0.5mM) was added to start the reaction, and the consumption of NADH at 456nm was monitored over time with a UV-Vis microplate reader. Protein activity was assessed using the initial reaction rate within 30 s, where N...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com