Bile salt hydrolase mutant with improved enzyme activity
A bile salt hydrolysis enzyme and mutant technology, applied in the field of enzyme engineering, can solve the problems of easy formation of inclusion bodies and low expression of bile salt hydrolysis enzyme, and achieve the effect of improving enzyme activity and production efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0020] Preparation of crude enzyme solution: Centrifuge the fermentation broth at 6000-9000rpm, collect the bacteria, wash the bacteria with 0.1M, pH 6-7 phosphate buffer 2-3 times, and ultrasonically break for 5min, then centrifuge to collect the supernatant That is the crude enzyme solution.
[0021] Bile salt hydrolysis enzyme activity assay method: Take 10 μL enzyme solution and dilute it to 90 μL with 0.1M phosphate buffer (pH 6.0), add 10 μL combined bile salt (200mM) and mix, incubate at 37°C for 30min, add an equal volume of 15% (w / v) trichloroacetic acid to terminate the reaction, after centrifugation, take 10 μL supernatant and mix with 190 μL ninhydrin reagent, react at 100 °C for 15 min, measure the absorbance at 570 nm after cooling, and calculate according to the glycine standard curve. Among them, the composition of ninhydrin reagent is: 0.5mL 1% (w / v) ninhydrin (dissolved in 0.5M, pH5.5 citrate buffer), 1.2mL glycerol, 0.2mL 0.5M, pH5.5 citrate buffer.
Embodiment 1
[0022] Example 1 Construction of Efficient Secretion of Bile Salt Hydrolase Strain
[0023] Using the amino acid shown in SEQ ID NO.1 as a template, the gene bsh shown in SEQ ID NO.2 was synthesized after codon optimization. The gene bsh1 was ligated with the vector pET-22b(+), the ligation system: 4 μL of the target gene bsh, 1 μL of the vector pET-22b(+), 5 μL of solutionI, ligated overnight at 16°C. Transform the ligated recombinant plasmid pET-22b-bsh1 into competent cells E.coil JM109, transform the ampicillin LB plate, pick positive colonies, inoculate them into LB medium, and extract the plasmid after overnight culture on a shaker at 37°C. After the enzyme digestion was verified to be correct, the transformants were sequenced by Shanghai Sangon.
Embodiment 2
[0024] Example 2 Verification of Bile Salt Hydrolase Production Strain with High Secretory Ability
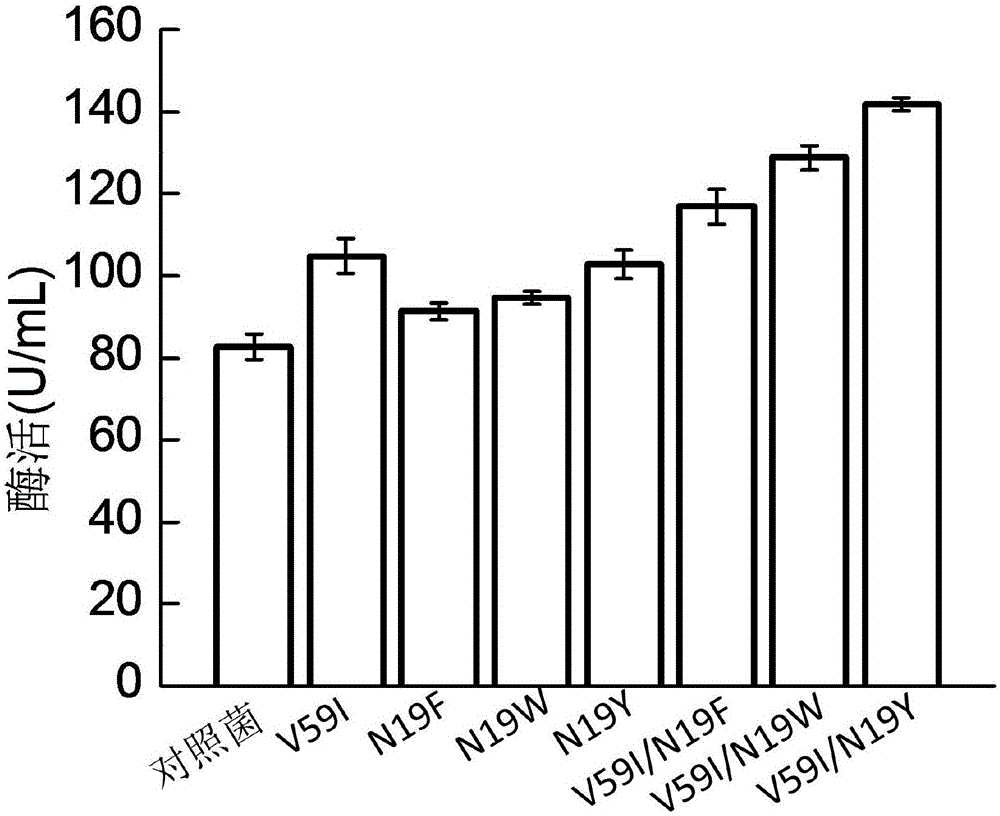
[0025] The plasmid sequenced correctly in Example 1 was transformed into Escherichia coli E.coli BL21(DE3). The selected transformants were inoculated into LB liquid medium, cultured at 37° C. for 12 hours, and then transferred into TB medium with an inoculation amount of 1%. Bacteria grow to OD 600 At 1.0, IPTG was added to induce, and the culture temperature was lowered to 28°C, and cultured for 36h. Centrifuge, collect the bacteria, and detect the intracellular enzyme activity. The results showed that the enzyme activity was 82.68U / mL.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com