Novel plant salt-resistant gene ZmDUF1644, and expression vector and application thereof
A plant expression vector, zmduf1644 technology, applied in the field of molecular biology, can solve the problems of low efficiency and long cycle, and achieve the effect of improving salt tolerance and plant salt tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
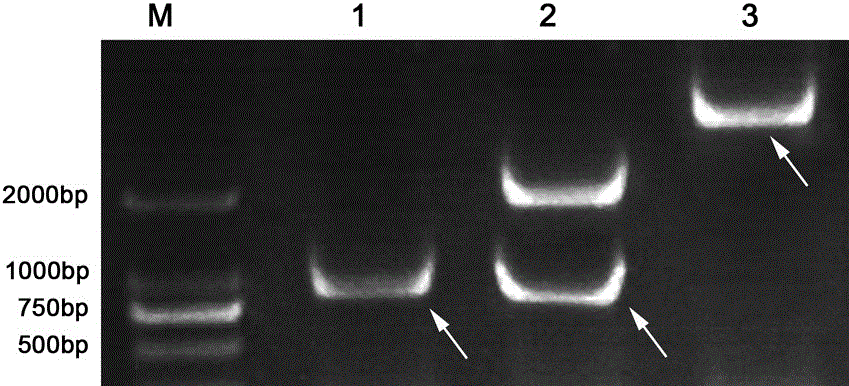
[0026] Example 1 Zoysia folium ZmDUF1644 clone.
[0027] Choose Zoysia ditch leaves ( Zoysia matrella ) as materials, select healthy turf pieces, place them in a cup filled with quartz sand, and culture them in 1 / 2 Hongland nutrient solution for 20 days, then transfer them to 1 / 2 Hongland nutrient solution containing 300mM NaCl for 7 days, take 0.1g of young leaves, according to the instructions of the Trizol RNA Extraction Kit (TaKaRa), extract the total RNA of the leaves, follow the M-MLV Reverse Transcription Kit (TaKaRa) to take 1 µg of total RNA and reverse transcribe into cDNA, and digest the cDNA with RNase products, designed primers to amplify ZmDUF1644 ;
[0028] Upstream primer ZmDUF1644-F: 5′-ATGCCAAAGGACAGGAGC-3′ (SEQ ID NO.2);
[0029] Downstream primer ZmDUF1644-R: 5′-TTGTGCAGGGTCACCGTC-3′ (SEQ ID NO.3).
[0030] Using the extracted leaf cDNA as a template, carry out PCR reaction, 20 µL reaction system: 10 µL of 2×LA RCR Mix, 1.0 µL each of ZmDUF1644-F and ...
Embodiment 2
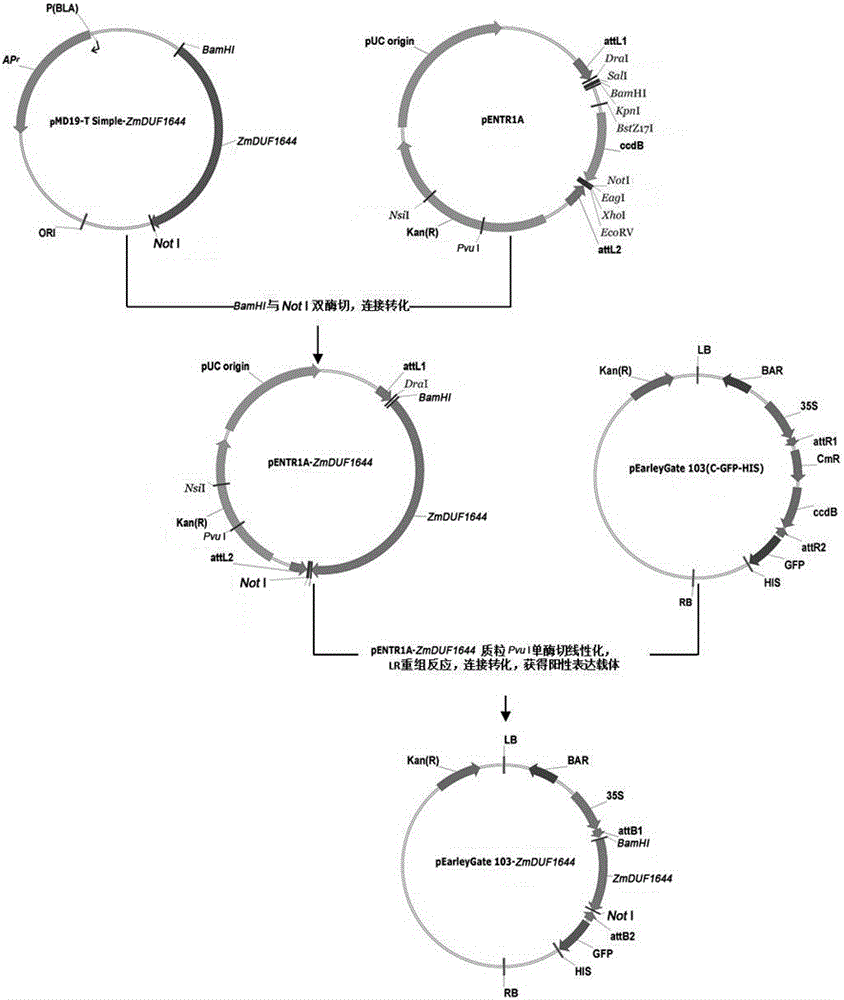
[0031] Example 2. Plant expression vector pEarleyGate103- ZmDUF1644 build.
[0032] Design primers for PCR reaction in target gene ZmDUF1644 Respectively introduce enzyme cutting sites upstream and downstream Bam H I and not Ⅰ. The PCR product was connected to the pMD19-T Simple vector, transformed into TOP10 competent cells, and the positive plasmid was extracted. Bam H I and not Ⅰ double digested ZmDUF1644 fragment with Bam H I and not Ⅰ The double-digested pENTR1A connection was transformed into Escherichia coli, and the extracted positive plasmid was Nsi ⅠAfter linearization by single enzyme digestion, carry out LR recombination reaction (Invitrogen) with pEarleyGate103 vector plasmid, transform, extract positive plasmid, electrophoresis detection and sequencing verification as SEQ ID NO.1;
[0033] Upstream primer ZmDUF1644-BamHI-F: 5′-GGATCCGGATGCCAAAGGACAGGAGC-3′ (SEQ ID NO.4);
[0034] Downstream primer ZmDUF1644-NotI-R: 5'-GCGGCCGCGATTGTGCAGGGTCACCGTC-3'...
Embodiment 3
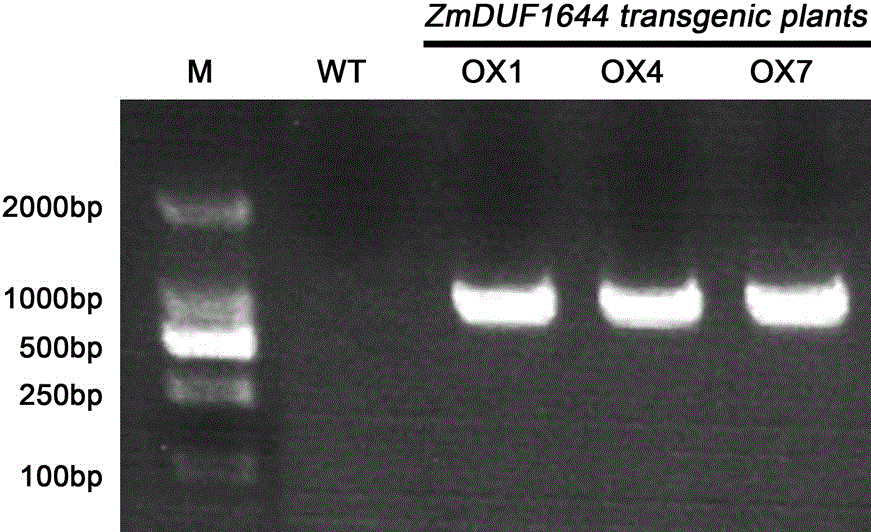
[0038] Example 3 Plant expression vector pEarleyGate103- ZmDUF1644 Genetic transformation of Arabidopsis thaliana and identification of its salt tolerance. ①Competent preparation of Agrobacterium strain EHA105 and transformation by freeze-thaw method: Pick a single colony of EHA105 from a YEB (50 µg / mL rifampicin) plate and inoculate it in 50 mL of YEB liquid medium containing 50 µg / mL rifampicin medium, 220 rpm, 28°C to OD value 0.6, then ice-bathed the bacteria solution for 30 min, centrifuged to collect the bacteria, suspended in 2 mL of pre-cooled 100mM CaCl 2 (20% glycerol) solution, 200 µL / tube for use; take 5 µL pEarleyGate103- ZmDUF1644 Add 100 µL of competent cells to the vector plasmid, ice bath for 30 min, freeze in liquid nitrogen for 5 min, 37°C for 5 min, add 800 µL of YEB liquid medium, pre-culture for 3 h at 28°C and 200 rpm, and smear the bacterial solution on YEB ( 50 µg / mL rifampicin + 50 µg / mL kanamycin) solid medium, cultured in the dark at 28°C for 2 d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com