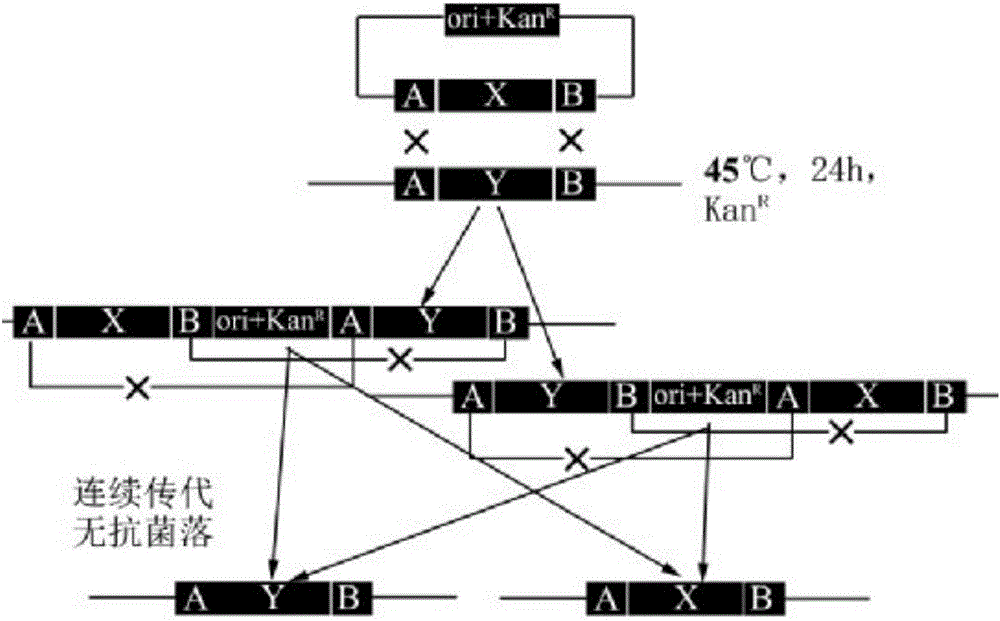
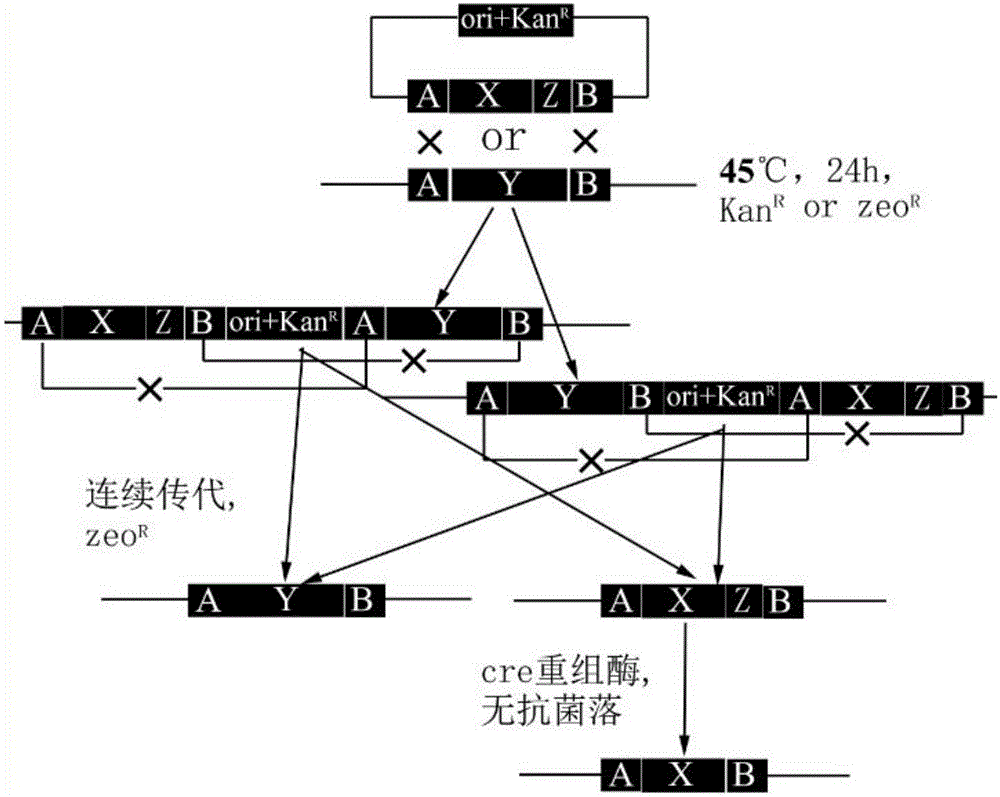
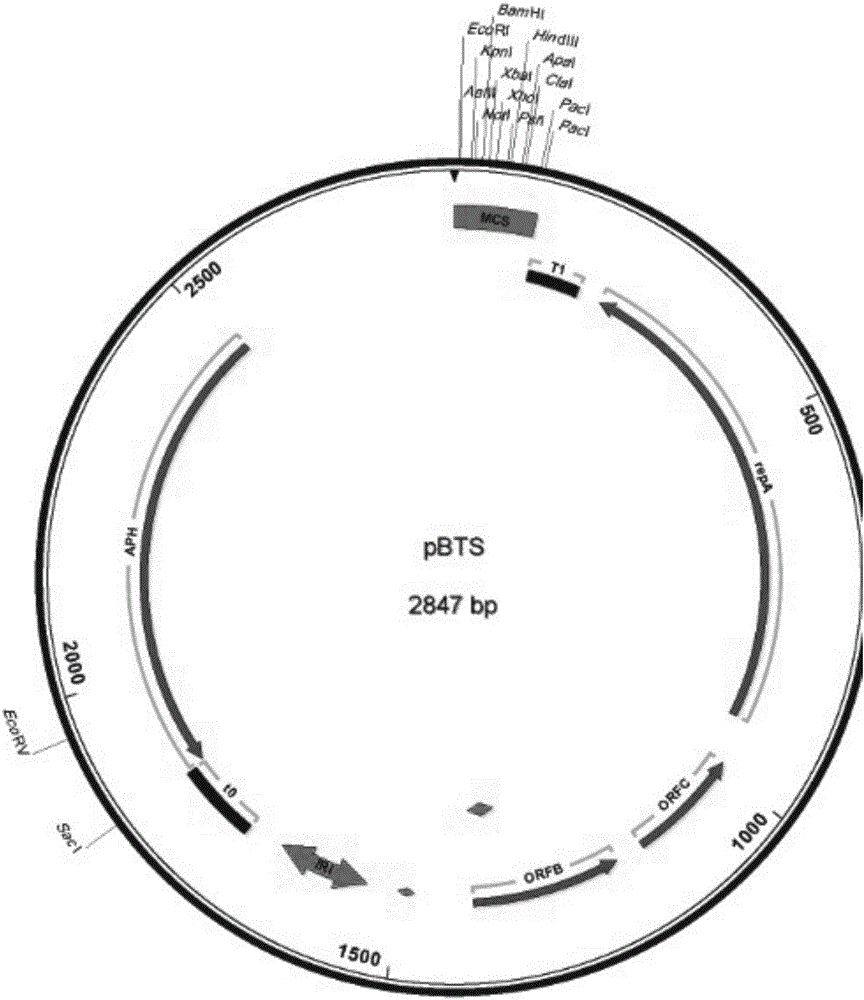
Method for transforming bacillus genome
A Bacillus and genome technology, applied in the directions of microorganism-based methods, biochemical equipment and methods, recombinant DNA technology, etc., can solve the problems of the research and increase of the temperature-sensitive characteristics of the modified plasmid pBAV1K, and achieve low difficulty and adaptability of plasmid transformation. wide effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] (1) Cut pBAV1K-T5-GFP plasmid with EcoR I and Apa I endonucleases, and perform homologous recombination with the synthesized MCS fragment to obtain plasmid pBAV1K.
[0048] The endonuclease used in this experiment and follow-up experiments was the rapid endonuclease from Thermo Company, and the gel recovery kit (DE-02011) from Chengdu Fuji Biotechnology Co., Ltd. was used for fragment recovery. The MCS fragment was synthesized by Jinweizhi Biotechnology Co., Ltd. Fragments and homologous recombination used Tiangen's EsayGeno rapid recombination cloning kit (VI201-02), the E. coli strain was top10, and the KCM method was used to prepare competent cells. DE-01001).
[0049] (2) Design primers to amplify the pBAV1K fragment with synonymous mutation to delete the Nde I restriction site, connect the fragments by homologous recombination cloning, and obtain a plasmid named pBTS.
[0050] The primers in this experiment and subsequent experiments were synthesized by Jinweizhi ...
Embodiment 2
[0052] The IC fragment was synthesized from the whole gene and inserted between XbaI and PstI of pBTS by homologous recombination to obtain the plasmid pBTS-IC. What is obtained here is the pBTS-IC plasmid in the preferred scheme; pBTS-FR-X can refer to the plasmid for transforming the xylAB gene below.
Embodiment 3
[0054] 1. Design primers to amplify the 493bp fragment xyl-F upstream of the xylose xylAB operon of the Z12 strain, and insert it between the EcoR I and BamHI sites of pBTS to obtain the plasmid pBTS-xyl-F; amplify the downstream 620bp fragment xyl-R , Insert between BamHI and Hind III of plasmid pBTS-xyl-F to obtain plasmid pBTS-xyl plasmid (knockout xylAB operon). The high-fidelity enzyme uses toyobo's KOD-Plus high-fidelity polymerase (KOD-201).
[0055] 2. Design primers to amplify the 693bp fragment upstream of the pgsBCA operon of the Z12 strain, insert it between the EcoR I and BamHI sites of pBTS, obtain a 558bp fragment downstream of the amplified plasmid pBTS-pgs-F, insert the BamHI and BamHI of the plasmid pBTS-pgs-F Between Hind III, plasmid pBTS-pgs was obtained. The zeo fragment was synthesized from the whole gene and inserted into the BamHI site of pBTS-pgs to obtain the plasmid pBTS-pgs-Z (the pgsBCA operon was knocked out, but the homologous recombination fra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com