Escherichia coli and Shigella detection primers, kits and detection methods based on specific sequences
A specific sequence, Escherichia coli technology, applied in the field of molecular biology, can solve the problems of inaccurate identification of species, low reproducibility rate, long experiment period, etc., and achieve good promotion and application value, high sensitivity and specificity, and species Consistent effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] The detection primers for Escherichia coli and Shigella based on specific sequences in this embodiment are shown in SEQ ID NO.1 and SEQ ID NO.2.
Embodiment 2
[0036]The E. coli and Shigella detection kits based on specific sequences in this example include: 2×Taq PCRMaster Mix (0.1U / μL Taq DNAPolymerase, 2×PCR reaction buffer, 0.4mM dNTP, 4mM MgSO 4 ) 20mL, sterilized ultrapure water 30mL, positive control DNA (Escherichia coli and Shigella each one) 100μL, 8μmol / L upstream and downstream primers 5mL each, DNA Marker DL 2000 (250μL) and kit instructions;
[0037] The upstream and downstream primers are:
[0038] Upstream primer: 5'-GTTATCAGCAACGCGCAAAA-3';
[0039] Downstream primer: 5'-ACTGGATGCGATGATGGATA-3'.
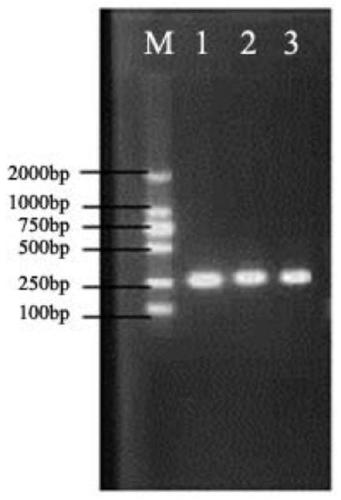
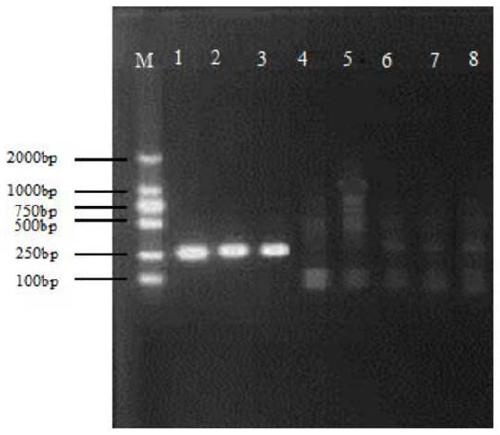
[0040] Detection principle: The kit contains various reagent components required for polymerase chain reaction, such as primers, dNTPs, buffer, Taq enzyme, etc., add detection samples to the PCR reaction solution, perform PCR amplification, and amplify the product The same band as the positive control was judged as suspected positive by electrophoresis analysis, the amplified product was recovered from the gel, and sequen...
Embodiment 3
[0050] The specific sequence-based detection method for Escherichia coli and Shigella in this embodiment comprises the following steps:
[0051] (1) Sample collection and pretreatment
[0052] Add 500 μL of the sample (food) to be tested to 10 mL of enrichment solution, enrich the bacteria at 37°C for 5 hours (4-6 hours are acceptable), centrifuge at 12000 rpm / min for 5 minutes in a desktop high-speed centrifuge, discard the supernatant, and use the precipitate as a detection template;
[0053] (2) PCR amplification
[0054] Use the upstream and downstream primers for PCR amplification, and the primers are as follows:
[0055] Upstream primer: 5'-GTTATCAGCAACGCGCAAAA-3',
[0056] Downstream primer: 5'-ACTGGATGCGATGATGGATA-3';
[0057] The reaction system for PCR amplification is: 2×Taq PCR Master Mix (0.1U / μL Taq DNA Polymerase, 2×PCR reaction buffer, 0.4mM dNTP, 4mM MgSO 4 ) 12.5 μL, 8 μmol / L upstream and downstream primers 1 μL each, enrichment pellet 2 μL, 1×PCR colony ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com