Amaryllidaceae plant lycoris aurea cytochrome P450 reductase 2 and coding gene and application thereof
A technology of cytochrome and reductase, applied in the fields of biotechnology and plant biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Embodiment 1, cloning of gene encoding cytochrome P450 reductase LaCPR2
[0054] The two primers synthesized respectively have the nucleotide sequences of SEQ ID NO: 3 and SEQ ID NO: 4 in the sequence listing.
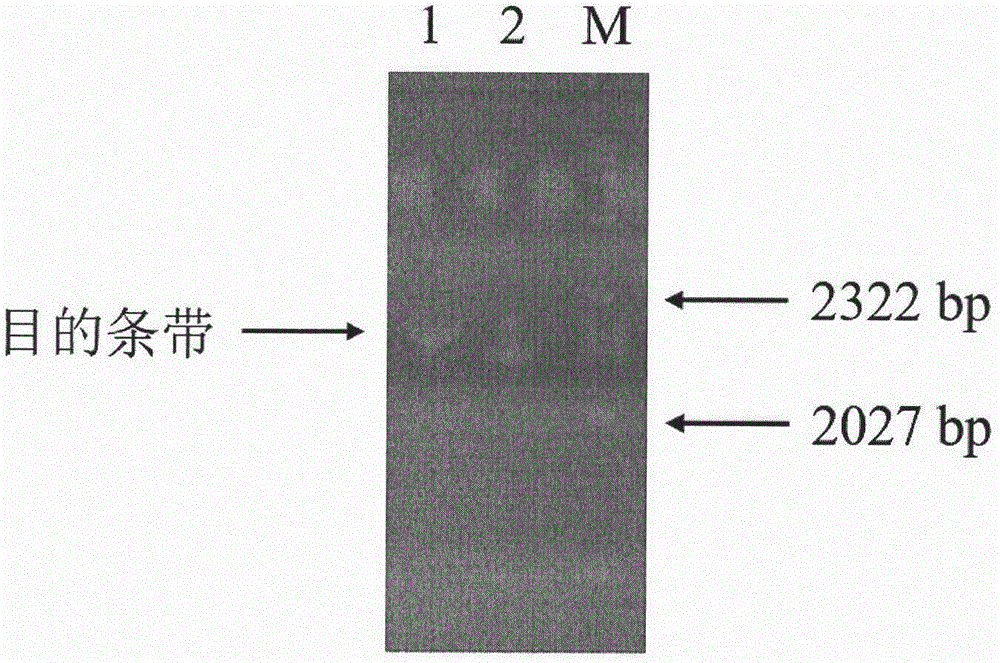
[0055] Using the cDNA obtained by reverse transcription of RNA extracted from Hudixiao as a template, PCR was performed using the above two primers SEQ ID NO: 3 and SEQ ID NO: 4. DNA polymerase was selected from Nanjing Nuoweizan Biotechnology Co., Ltd. Super-Fidelity DNA polymerase. The PCR amplification program was: 95°C for 5 min; 94°C for 45 s, 56°C for 45 s, 72°C for 2 min, a total of 30 cycles; 72°C for 10 min, then lowered to 10°C. The PCR products were detected by agarose gel electrophoresis, and the results were as follows: figure 1 .
[0056] Under the irradiation of ultraviolet light, cut out the target DNA band. Then, the DNA was recovered from the agarose gel using a multifunctional DNA purification kit (spin column type) (Beijing Biotech Biot...
Embodiment 2
[0058] The construction of the recombinant expression vector of embodiment 2, LaCPR2 gene
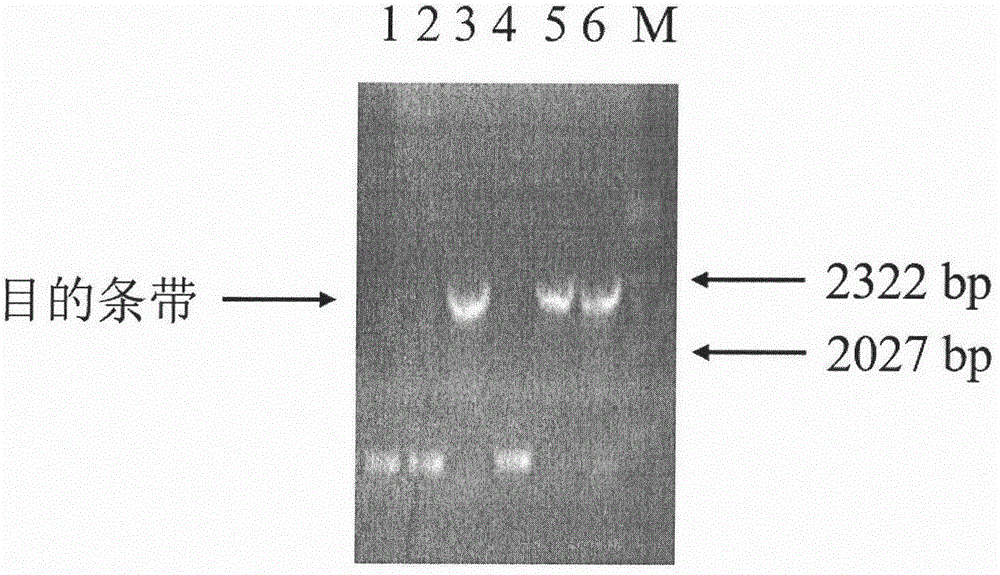
[0059] (1) Synthesize two primers respectively having the nucleotide sequences of SEQ ID NO: 5 and SEQ ID NO: 7 in the sequence listing. Two restriction sites, NdeI and XhoI, and their protected base sequences were respectively set at the 5'-ends of the synthesized primers SEQ ID NO: 5 and SEQ ID NO: 7, and PCR amplification was performed using the cDNA of Hudixiao as a template. The PCR amplification procedure is the same as in Example 1. The PCR amplified product was detected by agarose gel electrophoresis, separated, gel-cut and recovered, and then digested with NdeI and XhoI, and then ligated with T4 DNA ligase from Takara Bioengineering (Dalian) Co., Ltd. (TaKaRa) and also passed NdeI and XhoI double enzymes. cut pET28a vector (Novagen). The ligation product was transformed into Escherichia coli (E.coli) DH5α (purchased from Nanjing Novizan Biotechnology Co., Ltd.) competent cell...
Embodiment 3
[0061] Example 3, Expression, Purification and Enzyme Activity Determination of LaCPR2 and LaCPR2(ΔN66)
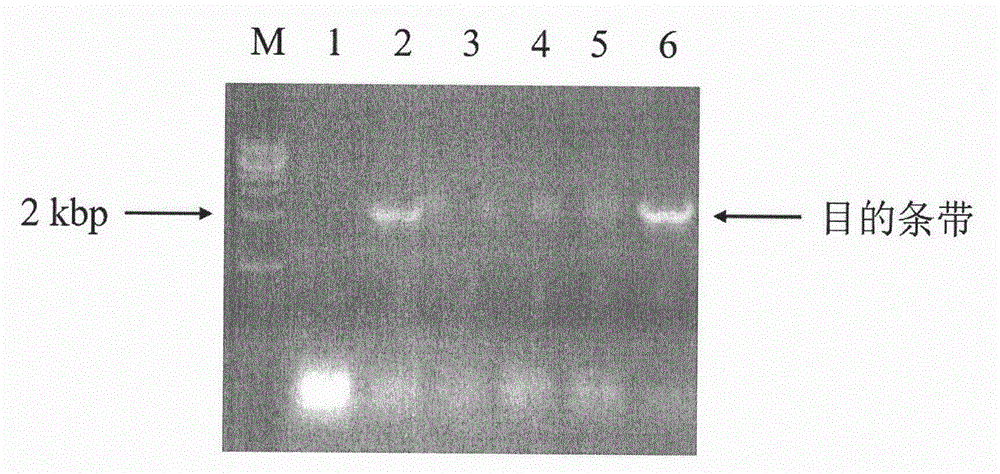
[0062] (1) The recombinant plasmid pET28a-LaCPR2 was transformed into Escherichia coli Rosseta(DE3) competent cells by heat shock method (42°C, 90s) to obtain recombinant Escherichia coli Rosseta(DE3) / pET28a-LaCPR2. Pick a single clone for overnight culture, and then dilute the bacterial solution 100 times in LB medium containing kanamycin (final concentration: 25 μg / mL). When the bacterial solution grows to an absorbance of 0.6-0.8 at a wavelength of 600 nm, the inducer isopropyl-β-D-thiogalactopyranoside (IPTG) (final concentration is 0.1 mmol / L) is added for induction culture, and the culture temperature is 25°C. Take 1ml of the induced bacterial solution, collect the bacterial cells by centrifugation (4000rpm, 10min, 4°C), discard the supernatant, wash the bacterial cells twice with ice-cold 1×PBS buffer, resuspend in 400μl 1×PBS buffer, Add 100 μl of 5×SDS Loading B...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Theoretical molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com