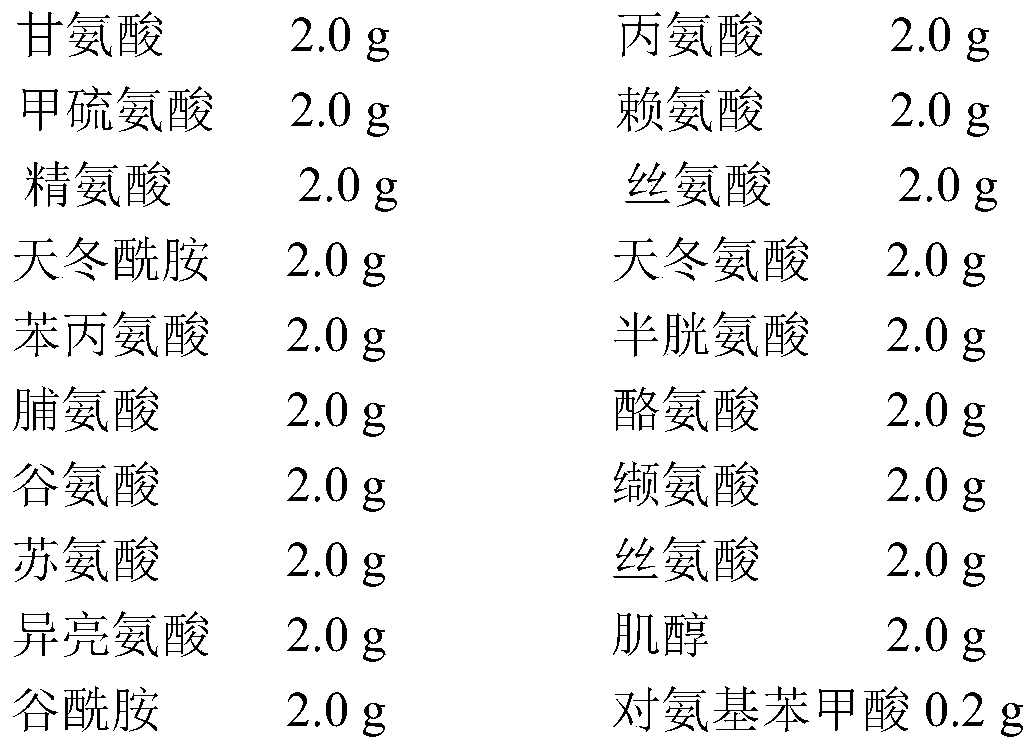
Recombinant saccharomyces cerevisiae for producing levopimaric diene and levopimaric acid and construction method
A technology for recombining Saccharomyces cerevisiae and L-pimamate acid, which is applied in the biological field to achieve the effect of increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1, source of each fragment and plasmid preparation method
[0026] (1) The gene element LPS sequence (levopimaradiene synthase LPS, GenPept: Q947C4.1) used in the present invention is derived from plant Ginkgo biloba (Ginkgo biloba); P450 s depend on cytochrome P450reductase CPR) is derived from the plant Taxus cuspidate, synthesized by Wuhan Jinkairui Bioengineering Co., Ltd. through chemical synthesis, and codon-optimized for Saccharomyces cerevisiae, and connected to the E. coli plasmid , preserved in E. coli. The nucleotide sequence of the optimized cytochrome P450 enzyme coding gene CYP720B1 is shown in SEQ ID NO.2 and the nucleotide sequence of the optimized cytochrome P450 reductase coding gene TcCPR is shown in SEQ ID NO.3.
[0027] (2) The method of transforming the gene LPS encoding L-pimarone diene synthase into TΔLPS of truncation mutation is as follows:
[0028] TΔLPS (the N-terminal truncation of 79 amino acids of the amino acid sequence encod...
Embodiment 2
[0054] Embodiment 2, the construction of Saccharomyces cerevisiae recombinant strain 1
[0055] Into Saccharomyces cerevisiaeW303-1a, USA ATCC208352, the transformed levopipine diene synthase encoding gene TΔLPS, the optimized cytochrome P450 enzyme encoding gene CYP720B1 and the optimized cytochrome P450 reductase encoding gene TcCPR were introduced to obtain the recombinant strain 1. The nucleotide sequence of the transformed levopipine diene synthase coding gene TΔLPS is shown in SEQ ID NO.1, and the nucleotide sequence of the optimized cytochrome P450 enzyme coding gene CYP720B1 is shown in SEQ ID NO.2 The nucleotide sequence of the optimized cytochrome P450 reductase coding gene TcCPR is shown in SEQ ID NO.3.
[0056] 1. Module construction
[0057] rDNA-up (SEQ ID NO.13), promoter TEF1 (SEQ ID NO.14), terminator ADH2 (SEQ ID NO.15), promoter PGK1 (SEQ ID NO.16), terminator ADH1 (SEQ ID NO. .17), promoter TDH3 (SEQ ID NO.18), terminator TDH2 (SEQ ID NO.19), rDNA-down (...
Embodiment 3
[0090] Embodiment 3, the construction of Saccharomyces cerevisiae recombinant strain 2
[0091] Introducing the 3-hydroxy-3-methylglutaryl-CoA reductase encoding gene tHMG1 into the recombinant bacterium 1 to obtain the recombinant bacterium 2, the 3-hydroxy-3-methylglutaryl-CoA reductase encoding gene tHMG1 The nucleotide sequence of is shown in SEQ ID NO.4.
[0092] 1. Preparation method of plasmid pRS304-tHMG1
[0093] According to Table 6, using the Saccharomyces cerevisiae genome as a template, ApaI-Tdh3p-F (SEQ ID NO.46) as a front primer, and Tdh3p-R-tHMG1 (SEQ ID NO.47) as a back primer, PCR amplified fragment P Tdh3 ; Using the Saccharomyces cerevisiae genome as a template, Tdh3p-tHMG1-F (SEQ ID NO.48) as the front primer, and tHMG1-R-Cyc1t (SEQ ID NO.49) as the back primer, PCR amplifies the fragment tHMG1, (encoding The amino acid sequence at the N-terminal of HMGR1 is truncated, and only the sequence of 503 amino acids encoding the C-terminal of the HMGR1 protein...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com