Recombinant bacterium for generating inosine, preparation method and application thereof
A technology of recombinant bacteria and inosine, which is applied in the field of microbial fermentation, can solve the problems of slow growth of strains and decreased production of inosine in strains, and achieve the effect of increasing inosine production and being suitable for popularization and application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
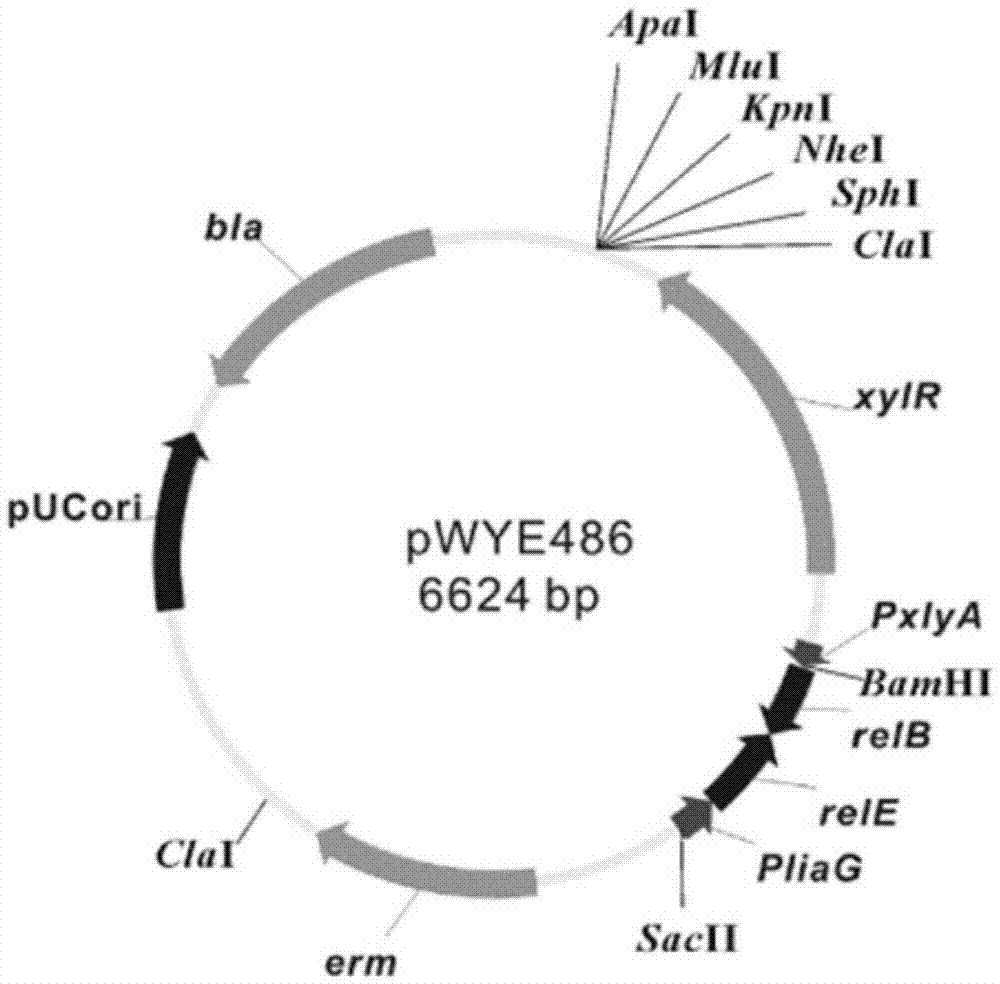
[0040] Example 1. Construction of the traceless genetic manipulation vector pWYE486 of Bacillus subtilis
[0041] (1) Construct carrying P liaG -relE-relB gene recombinant plasmid pWYE448
[0042] The genomic DNA of Bacillus subtilis W168 (purchased from Bacillus Genetic Stock Center, USA, catalog number 1A308) was used as template and P1 / P2 were used as primers to amplify P liaG Promoter gene (fragment 1); using Escherichia coli W3110 (purchased from the U.S. Coli Genetic Stock Center, item number 4474) genomic DNA as a template, P3 / P4 as primers to amplify the toxin protein relE gene (fragment 2); using Escherichia coli W3110 genome DNA is used as a template, and P5 / P6 is used as primers to amplify the antitoxin protein relB gene (fragment 3). After the PCR amplification was completed, the PCR products were recovered on agarose gel, and fragment 1, fragment 2, and fragment 3 were used as templates, and P1 / P6 were used as primers to carry out overlap extension polymerase ch...
Embodiment 2
[0064] Embodiment 2. Construction of inosine chassis engineering bacteria IR-1
[0065] In order to block the adenosine synthesis branch of the branch metabolic pathway of inosine synthesis, the first key enzyme adenylosuccinate synthetase (Adenylosuccinate synthetase, EC: 6.3.4.4) The coding gene purA gene is inactivated.
[0066] Using Bacillus subtilis W168 genomic DNA as a template, using P13 / P14 as primers to PCR amplify the upstream homology arm of the purA gene (nucleotide sequence shown in SEQ ID NO: 2), the size is 611bp; using P15 / P16 as primers Amplify the downstream homology arm of purA gene, the size is 650bp. After the PCR amplification was completed, the above two PCR products were recovered by agarose gel, and the recovered DNA product was used as a template and P13 / P16 as primers for SOE-PCR reaction to amplify to obtain purAUD. The resulting PCR fragment was recovered from agarose gel, digested with restriction endonucleases Kpn I and Sph I, and ligated wit...
Embodiment 3
[0073] Embodiment 3. Construction of inosine engineering bacteria IR-2
[0074] Pentose phosphate mutase catalyzes the reaction from (deoxy)ribose 1-phosphate to (deoxy)ribose 5-phosphate. In order to block the inosine decomposition pathway, the drm gene encoding pentose phosphate mutase (amino acid sequence shown in SEQ ID NO: 5) in the IR-1 strain was seamlessly knocked out (deleting its open reading frame).
[0075] Using Bacillus subtilis W168 genomic DNA as a template, using P17 / P18 as primers to PCR amplify the upstream homology arm of the drm gene (nucleotide sequence shown in SEQ ID NO: 3), the size is 700bp; using P19 / P20 as primers Amplify the downstream homology arm of drm gene, the size is 561bp. After PCR amplification, the above two PCR products were recovered by agarose gel, and the recovered DNA product was used as a template and P17 / P20 as primers for SOE-PCR reaction to amplify to obtain drmUD. The resulting PCR fragment was recovered from agarose gel, dige...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com