A gene encoding chloroplast carbonic anhydrase was used in the construction of high-concentration co 2 and applications in fast-growing industrial engineered microalgae
A technology of carbonic anhydrase and chloroplast, which is applied in the application field of industrial engineering microalgae, can solve the problems such as the lack of understanding of the CCM network regulation mechanism of Nannochloropsis and the lack of genetic transformation, so as to increase the growth rate, reduce production costs, and reduce pollution. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 Obtaining of the complete sequence of Nannochloropsis oceanica IMET1 carbonic anhydrase
[0033] By sequencing the whole genome of Nannochloropsis, the complete sequence of chloroplast carbonic anhydrase encoded by the nuclear genome was preliminarily obtained, as shown in SEQ ID No.1 and SEQ ID No.2; and the homologous sequence of SEQ ID No.2 SEQ ID No. 3.
[0034] The complete reading frame (ORF) of chloroplast carbonic anhydrase was further verified by polymerase chain reaction PCR. The PCR primer sequence was: F1 was 5'ATGTGGCGGCGTGTGCTCGCA3'; R1 was 5'CTAGACGCTTTTATTAACC3'. The PCR reaction system includes 5XPCR reaction buffer (5X DNA buffer) 5ul, deoxyribonucleoside triphosphate (dNTP) 4ul, magnesium sulfate (MgSO 4 ) 2ul, forward primer (Forward primer) 2ul, reverse primer (Reverse primer) 2ul, DNA 3ul, DNA polymerase (KOD DNA poleymase) 0.5ul, ddH 2 O is 32.5ul, and the total reaction system is 50ul;
[0035] The PCR reaction program was the first ...
Embodiment 2
[0037] 1) Construction of RNAi expression vector
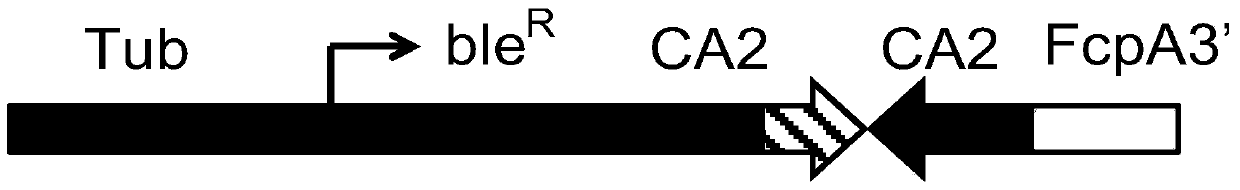
[0038] The construction of the RNAi expression vector uses phir-PtGUS as the carrier backbone. First, the Nannochloropsis genome is used as a template by PCR amplification reaction to amplify a 229bp fragment (CA2 gene sequence 1395bp to 1623bp position) and a 404bp fragment (CA2 gene sequence 1395bp to 1798bp position) two PCR fragments;
[0039] Wherein, the primers for amplifying the 229bp fragment are:
[0040] CA2_Fw (5'CGGAATTCGGGCATAGAGTGCGAATTGA3'; contains EcoRI site) / CA2_Rv1 (5'GCTCTAGACGCTTTTATTAACCCCATCC3'; contains XbaI site);
[0041] The primers for amplifying the 404bp fragment are:
[0042] CA2_Fw (5'CGGAATTCGGGCATAGAGTGCGAATTGA3'; contains an EcoRI site) / CA2_Rv2 (5'GCTCTAGAATCCTGGTCGTCAAAGAACG3'; contains an XbaI site).
[0043] The above reaction systems for PCR amplification are all 50ul: including 5XPCR reaction buffer (5X DNA buffer) 5ul, deoxyribonucleoside triphosphate (dNTP) 4ul, magnesium sulfate (...
Embodiment 3
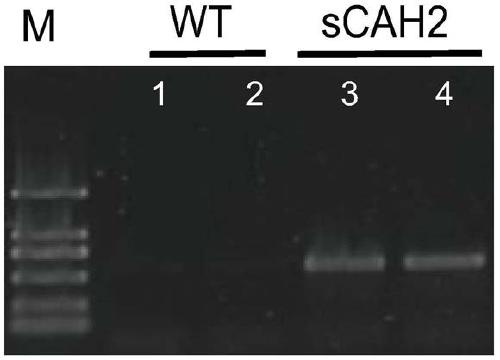
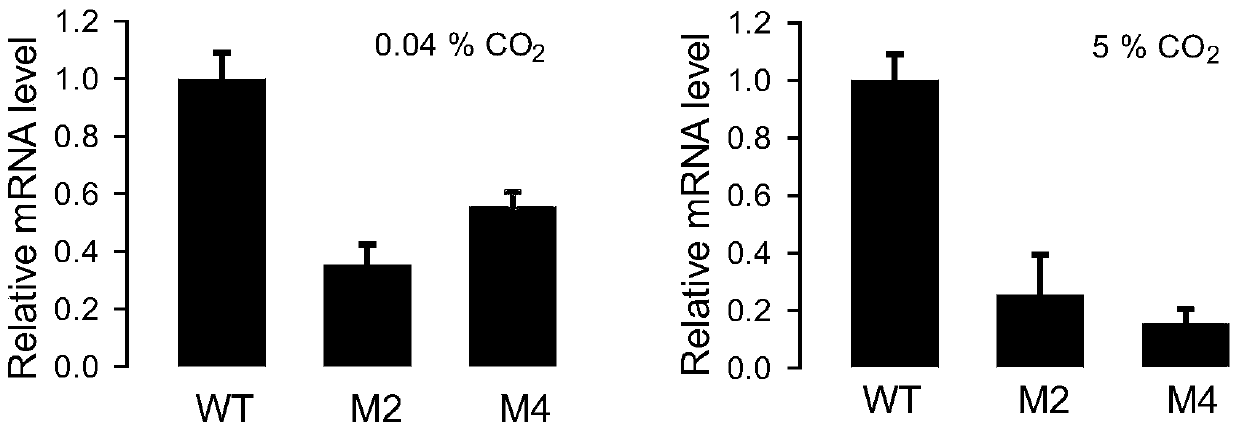
[0052] The PCR verification of embodiment 3 transformants
[0053] Pick the transformant with a toothpick and put it in fresh f / 2 medium (containing 3-5ug / ml Zeocin antibiotic), culture it in a light incubator, wait for it to grow for 2-3 weeks, collect the algae cells by centrifugation, and use genomic DNA extraction reagent The genomic DNA of the transformant was extracted using the OMEGA HP DNAextration kit, and the genomic DNA of the wild-type Nannochloropsis was also extracted as a control for subsequent experiments. After the genome was extracted and quantified by Nannodrop, PCR amplification was performed with forward primer F1 (5'TTATCAACGGCATACCGGCACTG3') and reverse primer R1 (5'CTGATGAACAGGGTCACGTCGT3'), the reaction system was 50ul: 5XPCR reaction buffer (5X DNA buffer) 5ul , deoxyribonucleoside triphosphate (dNTP) 4ul, magnesium sulfate (MgSO 4) 2ul, forward primer (primer F1) 2ul, reverse primer (primer R1) 2ul, template DNA (DNA) 3ul, DNA polymerase (KOD plus D...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com