miR156e-3p for regulating and controlling anthocyanin accumulation of Paeonia lactiflora Pall., plant expression vector of miR156e-3p and building method of plant expression vector
A mir156e-3p, plant expression vector technology, applied in the field of bioengineering, can solve problems such as unclear miRNA sequence, and achieve the effects of increasing anthocyanin content, increasing anthocyanin accumulation, and changing color.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
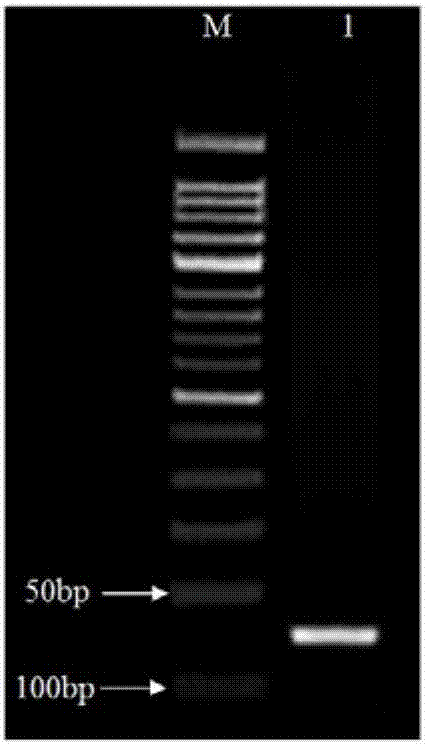
[0026] Cloning of Example 1.miR156e-3p Precursor Sequence
[0027] The red outer petals of peony 'Jinhui' were selected as the material, and the total RNA was extracted according to the instructions of the MiniBEST Plant RNA Extraction Kit (TaKaRa). TM RT reagent Kit with gDNAEraser kit (TaKaRa) took 1 μg of total RNA and reverse-transcribed it into cDNA.
[0028] Using the extracted petal cDNA as a template, design primers miR156e-3p-F and miR156e-3p-R for PCR reaction: upstream primer miR156e-3p-F: 5'-ACGAAGAAGAGAAAGAAATGTTGAC-3' (SEQ ID NO.3), downstream primer miR156e-3p-R: 5′-ACCCTTTAGCTGATCCCGGGTTGTG-3′ (SEQ ID NO.4); 25 μL reaction system: 10×RCR Buffer (Mg 2+ Plus) 2.5μL, miR156e-3p-F, miR156e-3p-R primers 1.25μL (10mM), dNTPmixture (2.5mM each) 2.0μL, Taq DNA Polymerase 0.25μL, cDNA template 2.0μL, ddH 2 O 15.75 μL; reaction program: pre-denaturation at 94°C for 3 min, then denaturation at 94°C for 30 sec, annealing at 56°C for 30 sec, extension at 72°C for 30 sec,...
Embodiment 2
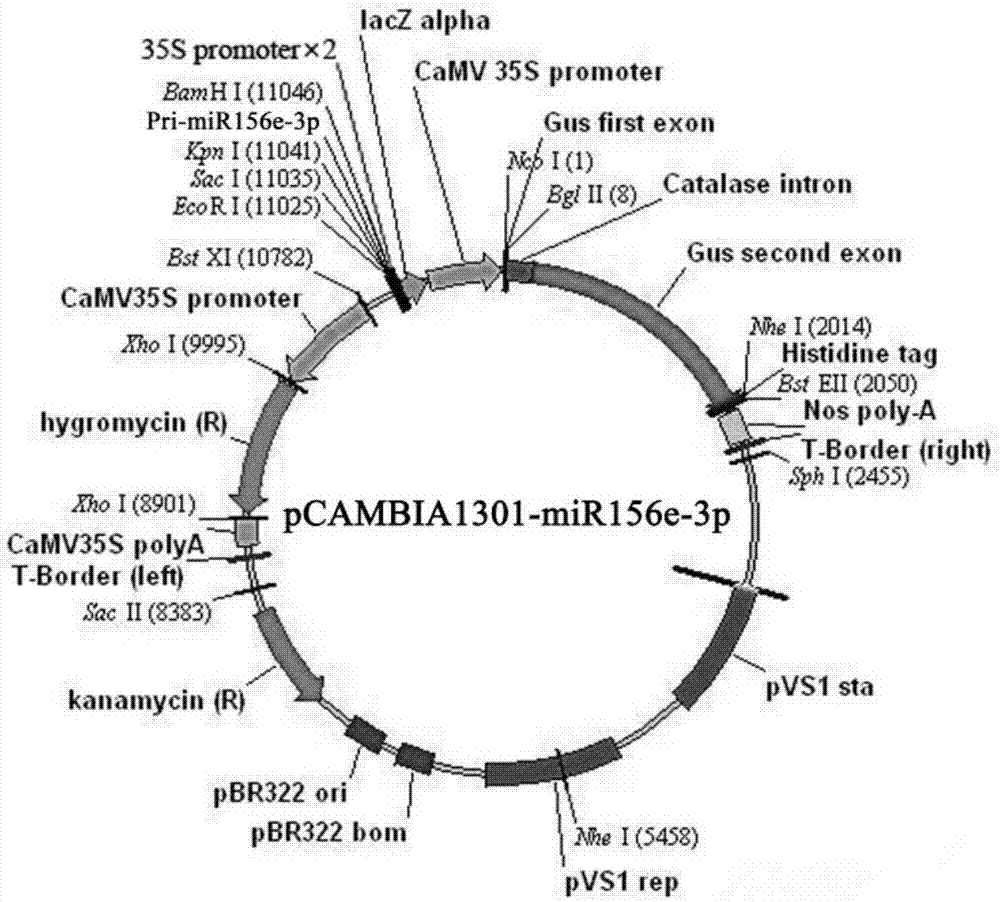
[0029] Example 2. Construction of plant expression vector pCAMBIA1301-miR156e-3p
[0030] Primers miR156e-3p-TF and miR156e-3p-TR were designed for PCR reaction, restriction sites BamH I and Kpn I were introduced upstream and downstream of the miR156e-3p precursor sequence respectively, and the PCR product was ligated into pEASY TM -T5 Zero CloningVector vector, transform Trans-T1 competent cells, then screen positive clones on LB plate with 0.1% ampicillin, extract positive plasmid, BamH I and Kpn I double-digested miR156e-3p precursor sequence fragment and BamH I and Kpn I double-digested pCAMBIA1301 was ligated, transformed, and the positive plasmid was extracted, detected by electrophoresis and sequenced to verify that it was SEQ ID NO.2. The specific steps were as follows: upstream primer miR156e-3p-TF: 5'-CGCGGATCCACGAAGAAGAGAAAGAAATGTTGAC-3'( SEQ ID NO.5), downstream primer miR156e-3p-TR: 5'-CGGGGTACCACCCCTTTAGCTGATCCCGGGTTGTG-3' (SEQ ID NO.6).
[0031] ①Using cDNA of ...
Embodiment 3
[0033] Example 3. Genetic transformation of Arabidopsis thaliana with plant expression vector pCAMBIA1301-miR156e-3p and evaluation of anthocyanin content and color
[0034] ① Competent preparation and freeze-thaw transformation of Agrobacterium strain EHA105
[0035] Pick a single colony of EHA105 from a YEB (50 μg / mL rifampicin) plate, inoculate it in 50 mL of YEB liquid medium containing 50 mg / L rifampicin, and culture it to OD at 28°C and 200 rpm 600 value 0.5, then ice-bathed the bacterial solution for 30 minutes, transferred to a pre-cooled 50mL centrifuge tube, collected the bacteria by centrifugation at 5000rpm at 4°C for 10 minutes, and suspended in 2 mL of pre-cooled 50mM CaCl 2 (15% glycerol) solution, after mixing, dispense 100 μL / tube into 1.5mL sterilized centrifuge tubes, freeze in liquid nitrogen and store at -80°C until use.
[0036] Take 10 μL pCAMBIA-miR156e-3p vector plasmid, add 200 μL EHA105 competent cells, bathe in ice for 30 minutes, freeze in liquid ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com