Multiple PCR detection primer groups for dairy cow mammitis pathogenic bacterium and application of multiple PCR detection primer groups
A technology for detecting primers for dairy cow mastitis, applied in the field of microorganisms, can solve the problems of low detection rate, unfavorable accurate detection of the bacteria and timely treatment of diseases, complicated operations, etc., so as to improve detection efficiency and protect milk and its dairy products. safe effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1: Construction of pMD-18T-pauA recombinant vector
[0038] According to GenBank published Streptococcus agalactiae sip gene (accession number HQ878436), Streptococcus dysgalactiae isp gene (accession number CP002215), Streptococcus uberis pauA gene (accession number KT006567.1), Staphylococcus aureus nuc gene (accession number DQ399678) designed primers to obtain: the upstream primer sip-F-2 and the downstream primer sip-R-2 for amplifying the Streptococcus agalactiae sip gene, the upstream primer isp-F-2 and the downstream primer for amplifying the Streptococcus agalactiae isp gene Primer isp-R-2, upstream primer pauA-F-2 and downstream primer pauA-R-2 for amplifying the pauA gene of Streptococcus uberis, upstream primer nuc-F-2 and downstream primer for amplifying the nuc gene of Staphylococcus aureus nuc-R-2; the nucleotide sequence of the primer is:
[0039]
[0040] Then, using genomic DNA as a template, the full-length sequences of sip, isp, pauA a...
Embodiment 2
[0041] Example 2: Specificity Evaluation Test
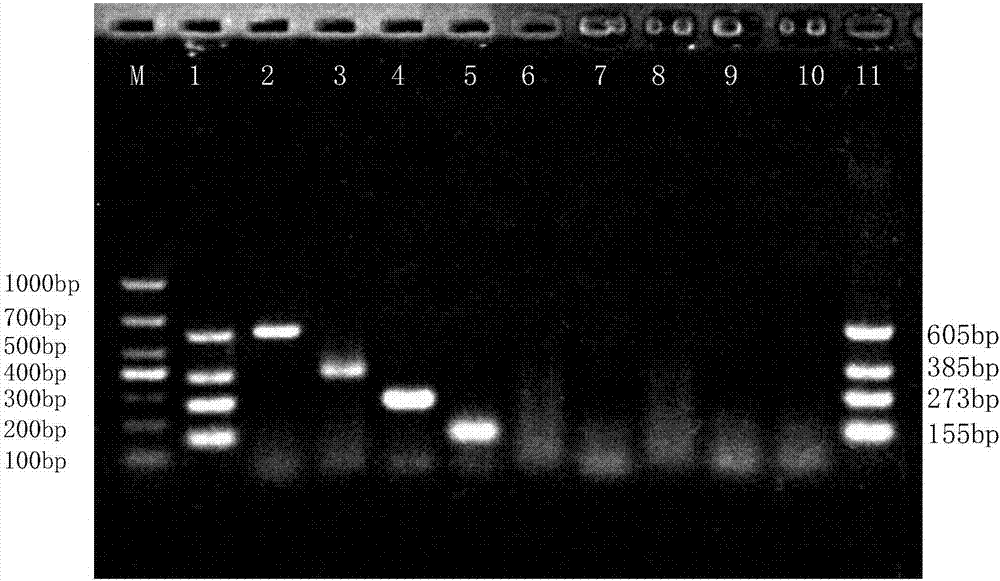
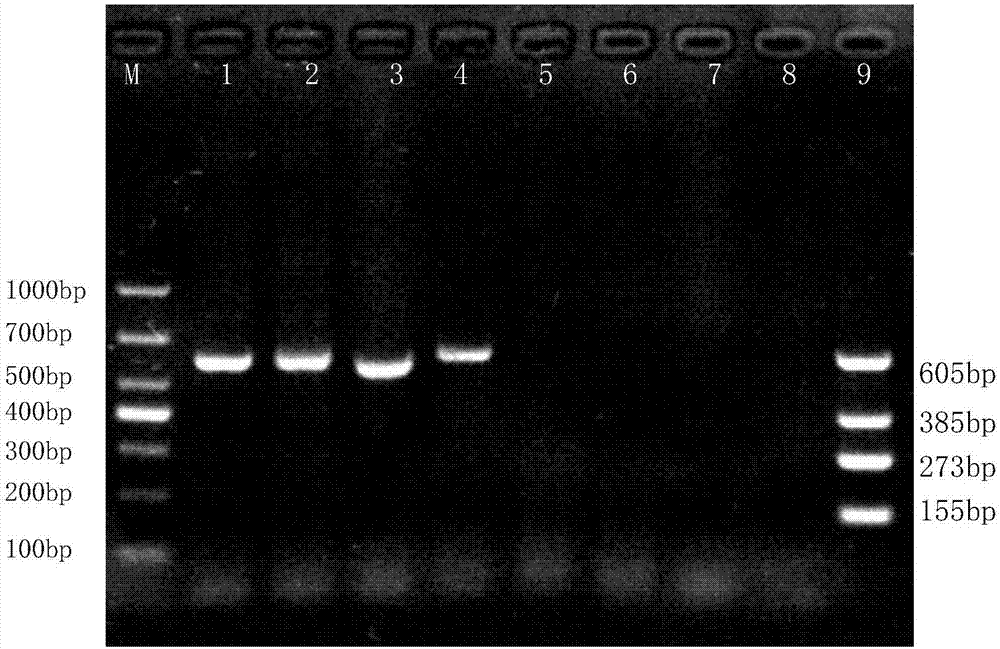
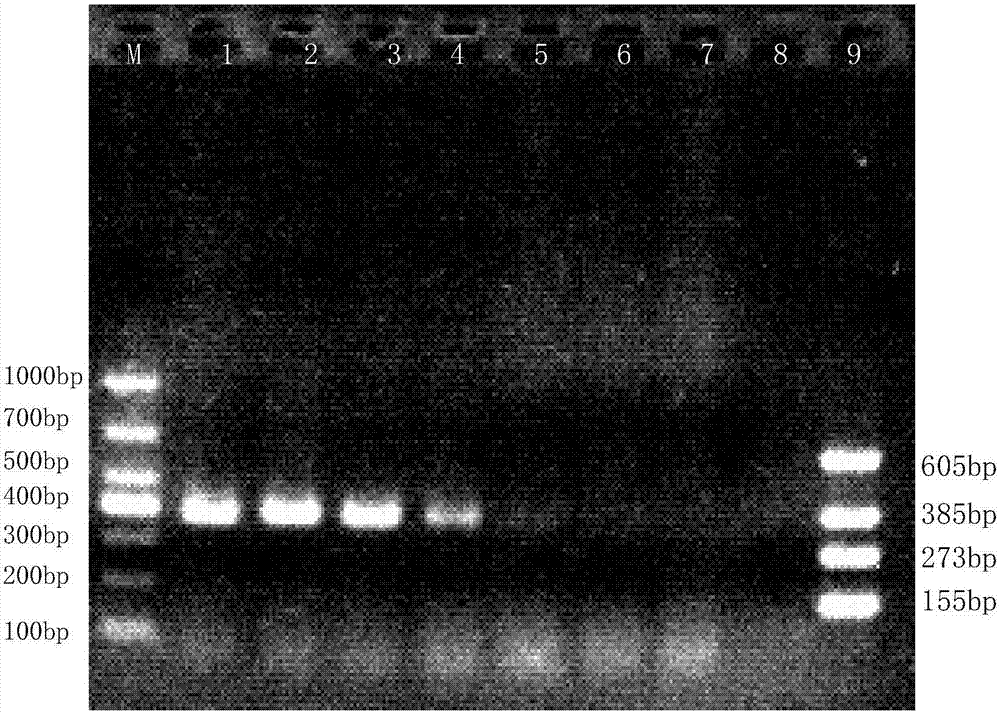
[0042] Perform PCR amplification reactions on Streptococcus agalactiae, Streptococcus dysgalactiae, Streptococcus uberis, Staphylococcus aureus, Escherichia coli, Salmonella, Streptococcus pneumoniae and Bacillus cereus isolated clinically and preserved by our laboratory,
[0043] The 50μL PCR detection system is:
[0044] Taq enzyme, 3.75U,
[0045] 10× PCR buffer, 5 μL,
[0046] MgCl 2 , the concentration is 1.5mmol / L,
[0047] dNTP, the concentration is 200μmol / L,
[0048] sip-F-1 and sip-R-1 primers, each with a concentration of 0.05 μmol / L,
[0049] isp-F-1 and isp-R-1 primers, the respective concentrations are 0.06 μmol / L,
[0050] pauA-F-1 and pauA-R-1 primers, each with a concentration of 0.16 μmol / L,
[0051] nuc-F-1 and nuc-R-1 primers, each with a concentration of 0.16 μmol / L,
[0052] Sample DNA template 50ng,
[0053] wxya 2 O to make up to 50 μL;
[0054] The conditions of PCR were: 95°C for 5 min; 35 cycle...
Embodiment 3
[0060] Example 3: Sensitivity Evaluation Test
[0061] Extract the recombinant vector constructed in Example 1, sterile ddH 2 O was diluted in a 10-fold gradient, and the copy number was taken as 10 8 、10 7 、10 6 、10 5 、10 4 、10 3 、10 2 The 7 concentration gradients are respectively templates, set sterile ddH 2 O is a negative control. The PCR reaction system is the same as in Example 2. 1.5% concentration of agarose gel electrophoresis to detect PCR amplification products.
[0062] The results of the susceptibility assessment test of Streptococcus agalactiae were as follows: figure 2 As shown, the lanes 1-7 in the figure are respectively: the copy number is 10 8 、10 7 、10 6 、10 5 、10 4 、10 3 、10 2 recombinant vector, lane 8 is sterile ddH 2 O negative control, lane 9 is positive control of four recombinant vectors, lane M: DL1,000 DNA Marker. It can be seen from the figure that the detection limit of PCR is 10 5 copy number.
[0063] For the results of t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com