Carbonyl reductase mutants and application thereof
A mutant, reductase technology, applied in the fields of genetic engineering and enzyme engineering, can solve problems such as large steric hindrance, and achieve the effects of improved space-time efficiency, good industrial application prospects, and a wide range of catalysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] The mensuration of embodiment 1 wild-type and mutant crude enzyme liquid enzyme activity
[0031] 1.1 Preparation of crude enzyme solution
[0032] Pick a single clone into LB (containing kanamycin 50 μg / mL) medium, culture overnight at 37 ° C, transfer to TB (containing kanamycin 50 μg / mL) medium with 1% inoculum size, 37 Cultivate at ℃ for 3h, add 0.5mM IPTG for induction, and continue to culture at 30°C for 18h. The bacterial solution was centrifuged at 4°C and 8000rpm to collect the bacteria, the cell homogenizer was crushed, and the supernatant was centrifuged to obtain a crude enzyme solution.
[0033] 1.2 Determination of crude enzyme activity
[0034] Crude enzyme activity assay reaction conditions: the reaction system consists of two phases, the aqueous phase is potassium phosphate buffer (0.1M, pH8.0) containing 3g / L crude enzyme solution (total protein concentration)), 0.2g / L NAD + ; The organic phase is isopropanol and substrate (substrate dissolved in i...
Embodiment 2
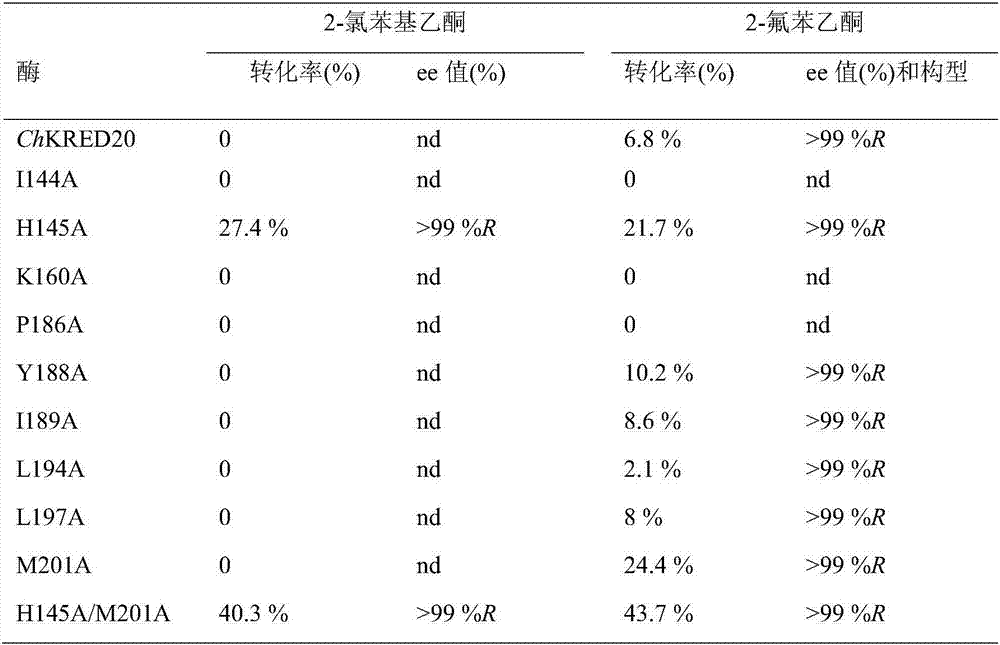
[0035] Example 2 Alanine scanning of the catalytic pocket of carbonyl reductase ChKRED20
[0036] 2.1 Construction method of alanine scanning mutants
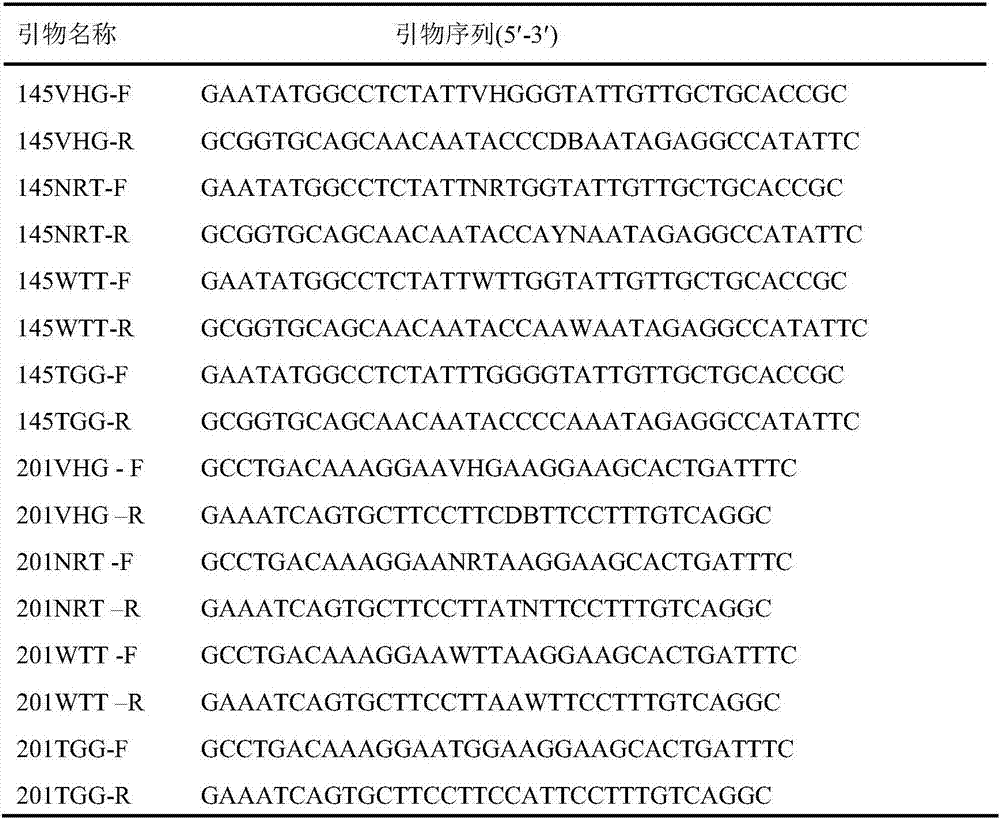
[0037] Based on the analysis of the crystal structure of the carbonyl reductase ChKRED20, we tried to perform alanine scanning on the 9 amino acid residues (I144, H145, K160, P186, Y188, I189, L194, L197, M201) in the catalytic pocket. Site-directed mutagenesis to alanine, all mutations were carried out using the carbonyl reductase ChKRED20 gene as a template, and the primers used are shown in Table 1.
[0038] Table 1 Primer sequences for alanine scanning
[0039]
[0040] The PCR conditions are: 10×Buffer 5 μL, each primer (10 mM) 6 μL, dNTP (2.5 mM) 4 μL, pfu enzyme (2.5U / mL) 1 μL, plasmid 10 ng, and ultrapure water to make up 50 μL. Conditions: Pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 68°C for 6 min, a total of 16 cycles. The PCR product was treated wi...
Embodiment 3
[0052] Embodiment 3 distance from the 145th histidine site randomization within
[0053] 3.1 Distance from the 145th histidine Saturation mutagenesis library construction method within
[0054] Saturation mutations were performed on the 143rd, 144th, 151st, 153rd, 160th, 186th, 188th, 196th, and 205th positions on the mutant H145A / M201A, respectively. Using mutant H145A / M201A as a template, NNS degenerate primers (N represents A, T, C, G; S represents G, C) are used. The degenerate primers used are shown in Table 4.
[0055] Table 4 The first round of single-site saturation mutation degenerate primers
[0056]
[0057] The PCR conditions are: 10×Buffer 5 μL, each primer (10 mM) 6 μL, dNTP (2.5 mM) 4 μL, pfu enzyme (2.5U / mL) 1 μL, plasmid 10 ng, and ultrapure water to make up 50 μL. Conditions: Pre-denaturation at 95°C for 5 min, denaturation at 95°C for 30 s, annealing at 55°C for 30 s, extension at 68°C for 6 min, a total of 16 cycles. The PCR product was treated wi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com