NGS database creating primer pool used for amplifying multiple targets in cfDNA sample and application
A primer pool and universal primer technology, applied in DNA/RNA fragmentation, recombinant DNA technology, microbial determination/inspection, etc., can solve the problem of difficult implementation of multiplex PCR technology, and achieve low non-specific amplification and easy amplification. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
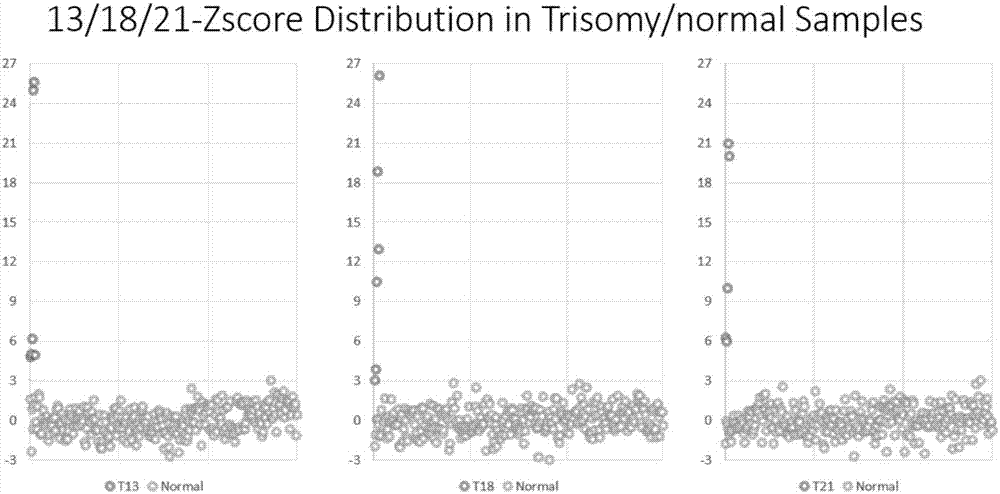
[0053] Example 1: Detection of fetal chromosomal aneuploidy using peripheral blood of pregnant women. It is used to verify the application of the present invention in the field of NIPT (non-invasive prenatal testing), which can correctly identify multiple chromosomal aneuploidies selected from trisomy 21, trisomy 13, trisomy 18 and monosomy X positive sample.
[0054] 1. Blood sample collection and processing
[0055] 1) 5ml of peripheral blood was collected intravenously from 275 pregnant women (gestational weeks over 12 weeks).
[0056] 2) Plasma separation: the blood collection tube was centrifuged at 1600 g for 10 min. Take supernatant plasma and transfer to EP tube. Centrifuge the above-mentioned EP tube at 16000 g for 10 min, point the tip of the pipette to the non-white blood cell precipitate, and absorb the supernatant. Proceed to the next step immediately or store in a -80°C refrigerator.
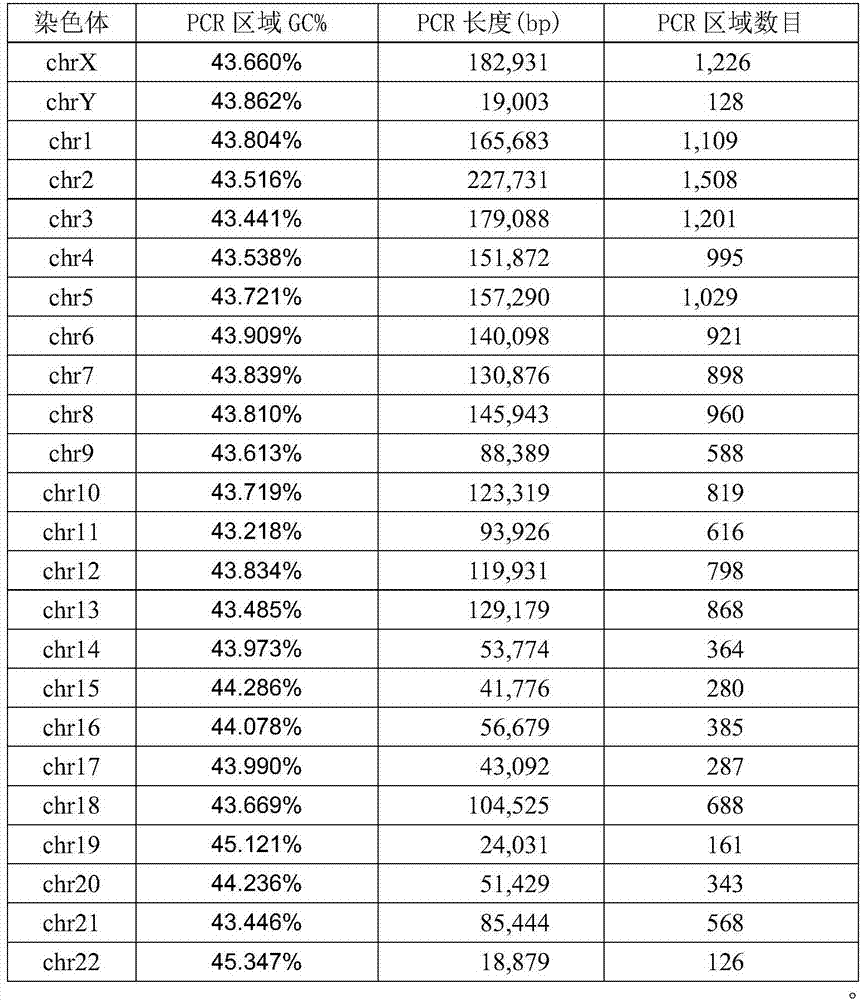
[0057] 3) Use Qiagen plasma DNA extraction kit to extract cfDNA in plasma...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com