SNP molecular marker related to reproductive characters of Chinese Holstein cow and applications of SNP molecular marker
A molecular marker and molecular marker-assisted technology, applied in the field of molecular biology, can solve the problems of disturbing the body's energy homeostasis, insulin sensitivity, affecting the transcriptional activity and expression of PPARγ, binding and covalent modification activation, etc., to accelerate genetic progress. , The method is accurate and reliable, and the detection cost is low.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 Determination of SNP markers related to reproductive traits of Chinese Holstein dairy cows and establishment of a method for detecting the loci
[0036] 1. Extract the genomic DNA from the blood of Chinese Holstein cows to be tested
[0037] Blood samples from 293 Chinese Holstein cows in different pastures were collected from the jugular vein, anticoagulated with ACD, and stored at -20°C for later use. The genomic DNA in bovine blood exists in the solution in the centrifuge tube, which can be stored at 4°C for later use or at -20°C for long-term storage.
[0038] 2. Amplify the nucleotide fragment containing the SNP site
[0039] Primers were designed according to the sequence of the PPARG gene (GenBank No.NC_007320.6) included in the NCBI database, including forward primer F: 5′-TGTAACTCAACCTCCTGTT-3′ and reverse primer R: 5′-AAGATGCTGTCAGTGAACT-3′, using the above genomic DNA As a template, the nucleotide fragment where the SNP to be detected is located i...
Embodiment 2
[0047] Example 2 Correlation analysis and detection application of different genotypes and reproductive traits of Chinese Holstein cows
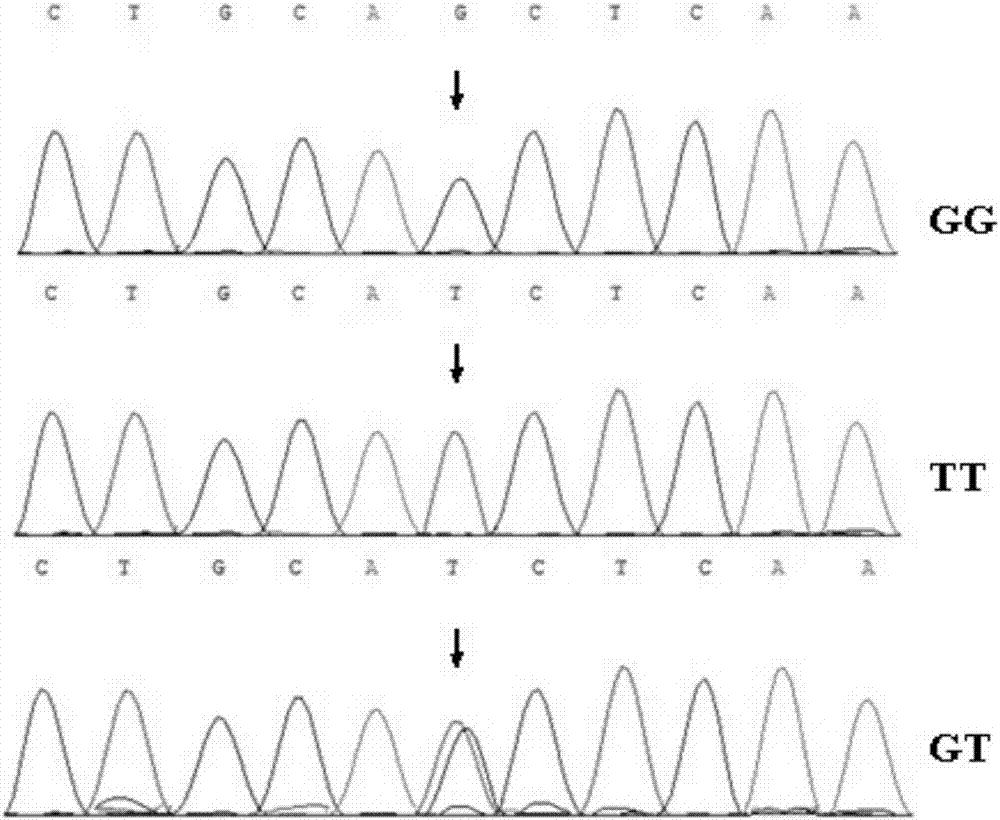
[0048] According to the method of Example 1, 293 Chinese Holstein dairy cows were detected by PCR-SSCP, and the analysis results of the 118bp site of the sequence shown in SEQ ID NO.1 of the PPARG gene are shown in Table 1.
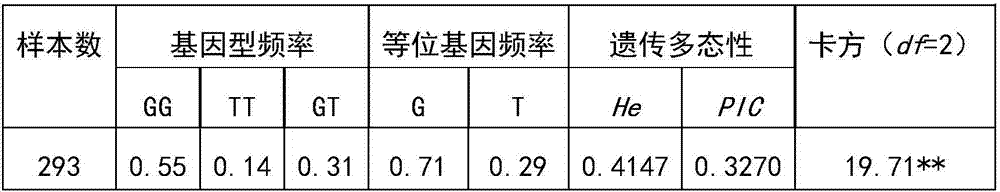
[0049] Table 1 Genetic polymorphism analysis of SNP loci in Chinese Holstein cow population
[0050]
[0051] It can be seen from Table 1 that GG individuals have the highest frequency in the test population, and G is the dominant allele. He was 0.4147, PIC was 0.3270, and was moderately polymorphic. χ obtained by Pearson chi-square test 2 19.71>χ0.01 2 (df=2)=9.21, then P<0.01, the test population deviates from the Hardy-Weinberg equilibrium state very significantly at this site, indicating that its mutation is affected by factors such as drift, selection and introduction.
[0052] Use SPSS software to call the ...
Embodiment 3
[0068] Example 3 Assembly and application of a kit for identifying Chinese Holstein cows with high reproductive performance
[0069] The components in the kit are as follows: 100ng / μL bovine blood DNA; upstream and downstream primers, the sequences of which are shown in SEQ ID NO.2-3 respectively, 2.5mmol / L dNTPs; 2.5U / μL Taq DNA polymerase; 10×Taq Buffer (Containing 15mmol / LMgCl 2 );Ultra-pure water.
[0070] F: 5'-TGTAACTCAACCTCCTGTT-3' (SEQ ID NO.2)
[0071] R: 5'-AAGATGCTGTCAGTGAACT-3' (SEQ ID NO.3)
[0072] The total PCR reaction system in the kit is 25μL, including 1μL of genomic DNA, 10×buffer (15mmol / LMgCl 2 ) 2.5μL, dNTP (2.5mmol / L) 2μL, upstream and downstream primers (12.5pmol / μL) each 0.5μL, Taq DNA polymerase (2.5U / μL) 0.5μL, ddH 2 O 18.0 μL. PCR reaction program: pre-denaturation at 95°C for 3min; denaturation at 94°C for 30s, annealing at 58°C for 30s, extension at 72°C for 45s, 35 cycles; final extension at 72°C for 10min.
[0073] The product obtained fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com