Kit for detecting achondroplasia (ACH) and hypochondroplasia (HCH) of neonates
A reagent and fetal technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., to achieve high detection sensitivity, high accuracy, and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1: Primer design and synthesis.
[0081] For the four mutation types at bases 1108, 1138 and 1620 of the FGFR3 gene, the corresponding specific PCR primer core sequences (SEQ ID No: 1 to SEQ ID No: 6) and specific extension primer core sequences (SEQ ID No: 7 to SEQ ID No: 9).
[0082] Among them, in order to prevent the PCR primers from entering the detection window of the mass spectrometer and interfering with the detection effect, a certain number of bases can be added to the core sequence (SEQ ID No: 1 to SEQ ID No: 6) at the 5' end of each PCR primer , common such as 10bp tag (ACGTTGGATG), to increase the molecular weight of the PCR primer, thereby exceeding the detection window of the mass spectrometer.
[0083] Related primers were synthesized at Sangon Bioengineering (Shanghai) Co., Ltd.
[0084] The following detection process is performed with reference to the "Folic Acid Genetic Metabolism Gene Mutation Detection Kit (Time-of-Flight Mass Spectrometr...
Embodiment 2
[0085] Example 2: Extraction of Free DNA from Peripheral Plasma of Pregnant Women
[0086] The blood sheet used is a circular filter paper with a diameter of 2 cm. Take 50ul of whole blood, drop it on the center of the circular filter paper, and then dry it for later use. The specific steps of blood sheet DNA extraction are as follows:
[0087] (1) Prepare the scissors and pre-treat them with 75% alcohol. Then cut out the circular piece of paper with blood spots and cut it into small pieces, put it into a 1.5ml centrifuge tube, add 20ul proteinase K, 20ulDTT and 500ul Lysis Buffer 1, mix well (vortex) , placed at 56°C for 2h (during which the mixture was inverted and mixed 3-4 times).
[0088] (2) Take out the centrifuge tube, centrifuge briefly, and use a pipette to transfer the supernatant as completely as possible to a new 1.5ml centrifuge tube to remove the interference of the filter paper on the next step of the experiment.
[0089] (3) Add 850ul Binding Buffer 2 and 1...
Embodiment 3
[0098] Embodiment three: Biological experiment.
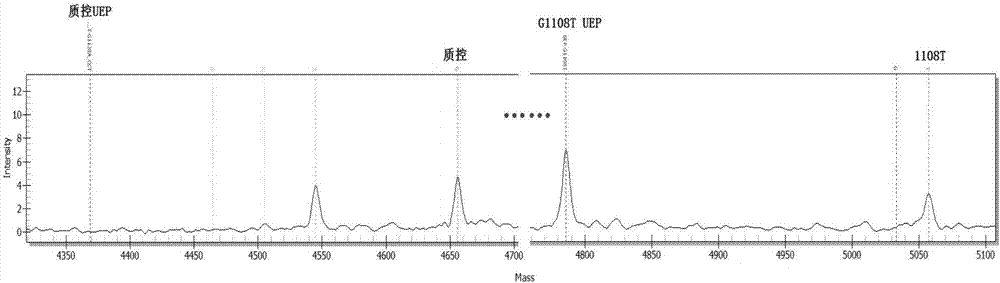
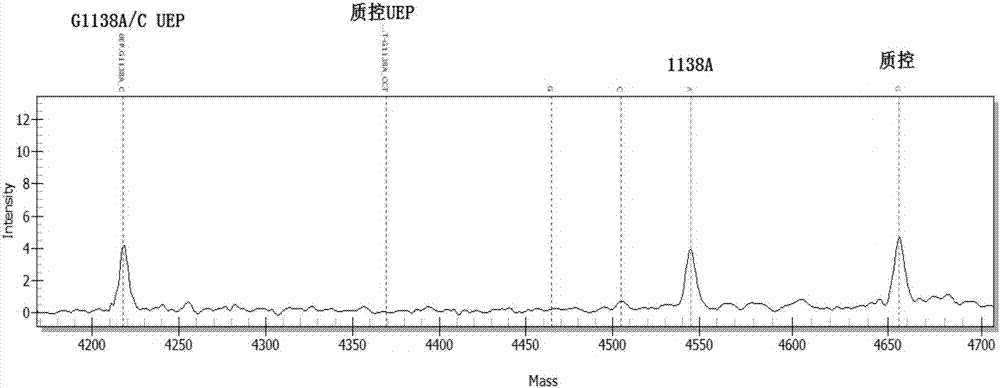
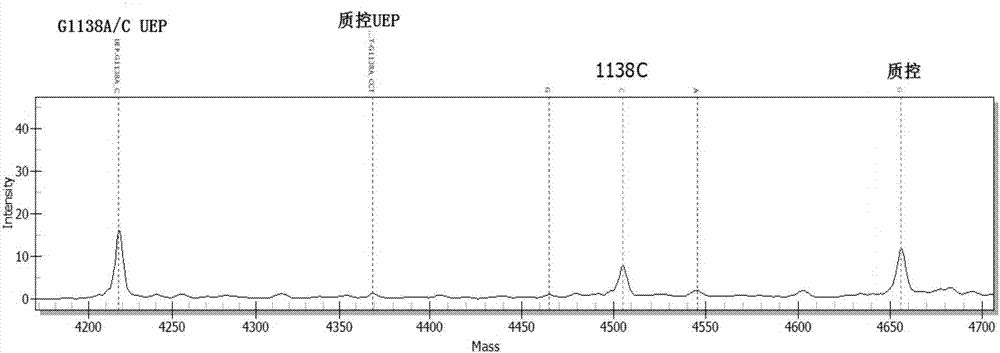
[0099] Using ABI 9700 PCR instrument, according to the instruction manual, the 3 target detection sites of FGFR3 gene were tested.
[0100] The components used in the kit for PCR, PCR product purification and single base extension are shown in Table 5:
[0101] table 5
[0102]
[0103] According to the manual, the specific operation method is as follows:
[0104] 1. PCR amplification
[0105] 1.1 In the PCR preparation area, prepare 200ul PCR reaction tubes according to the number of samples to be tested (including positive quality control products, negative controls, and blank controls), and mark the sample number on the tube;
[0106] 1.2 Take out the PCR mixture and PCR enzyme from the kit, let it thaw naturally, vortex and oscillate to mix well, and centrifuge instantly to the bottom of the tube;
[0107] 1.3 According to the number of samples, take out the PCR primer mixture and PCR reaction solution according to t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com