Quantitative detection method of mutant gene
A quantitative detection method and mutation gene technology, applied in the field of high-fidelity deep sequencing detection, can solve the problems of insufficient sample volume, high cost of digital PCR, large amount of DNA, etc., achieve high sensitivity and avoid the effect of low connection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Detection of Codon 12 and Codon 13 Mutations of Kras Gene in Peripheral Blood Plasma
[0045] 1. Sample DNA extraction
[0046] An appropriate amount of peripheral blood plasma from tumor patients was taken, and the cell-free genomic DNA was extracted with a cell-free DNA extraction kit (DK607-01, provided by Shanghai Laifeng Biotechnology Co., Ltd.).
[0047] 2. Level 1 Targeted Synthesis Amplification
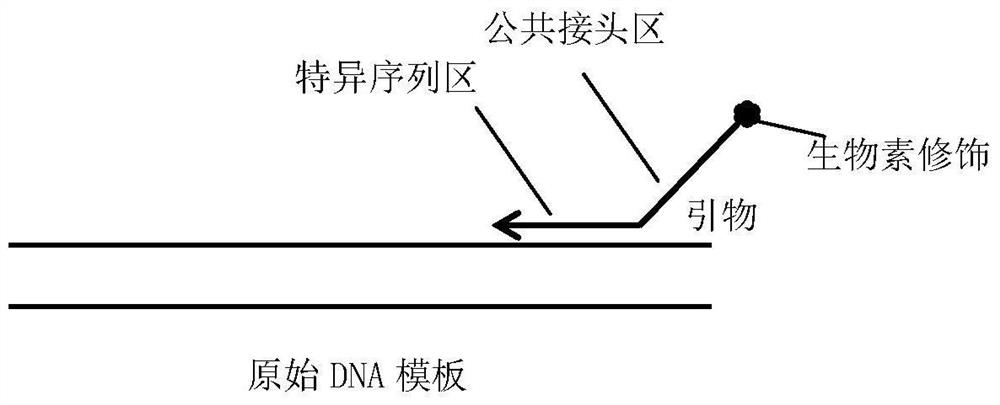
[0048] Reverse primer K-ras-41bp-R: T (biotin modified) TGCTCTTCCGATC-GTTCGTCCACAAAATGATTCTGA (SEQ ID NO: 1)
[0049] The reaction system of primary targeted synthesis amplification PCR is: 2 μL of 5×PCR buffer, 0.5 μL of 10 mM dNTP, 1 μL of DNA template, 1 μL of 5 μM reverse primer, 0.2 μL of 5 U / μl KOD DNA polymerase, H 2 O 5.8 μL, total volume 10 μL.
[0050] Reaction conditions are as shown in table 1:
[0051] Table 1 Reaction conditions for primary targeted synthesis amplification PCR
[0052]
[0053]
[0054] 3. Single-stranded product capt...
Embodiment 2
[0100] Example 2 Detection of hot zone mutations in peripheral blood plasma polygenes (Kras, Pten)
[0101] Detection method and condition are basically the same as in Example 1, except that:
[0102] In step 2, in the first-level targeted synthesis amplification reaction, in addition to the reverse primer K-ras-41bp-R, the primers used also have a reverse primer Pten-46bp-R: T (biotin modified) TGCTCTTCCGATC-AGTATCGGTTGGCTTTGTCT( SEQ ID NO: 12);
[0103] In step 4, in the protective excision purification reaction of the 3' end of the primary product, in addition to adding the sequence shown in SEQ ID NO: 2, the sequence: CGTGCAGATAATGACAAGGAAT (SEQ ID NO: 13);
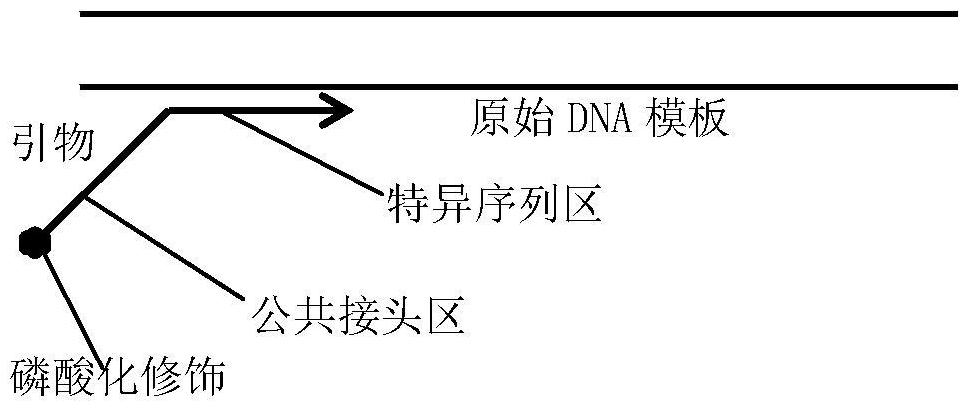
[0104] In step 5, in the secondary single-strand synthesis amplification reaction, the primers used, in addition to the forward primer K-ras-41bp-F, also have a forward primer Pten-46bp-F: C (phosphorylated modification) GCTCTTCCGATCT- CGTGCAGATAATGACAAGGAAT (SEQ ID NO: 14);
[0105] In step 6, in the protective ex...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com