Polystyrene affinity peptide and method of polystyrene affinity peptide for improving immobilized effect of antigen
A polystyrene, affinity technology, applied in the field of affinity peptide sequence PB-TUP, can solve the problems of loss of function, affect detection sensitivity and accuracy, loss of biological activity, etc., to avoid inactivation, improve immobilization effect, Effect of improving detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
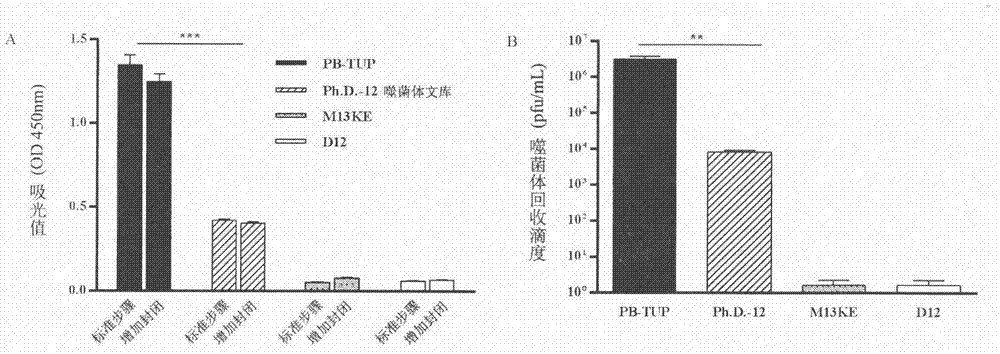
Embodiment 1
[0034] Screening for sequences with specific affinity to polystyrene microplates
[0035] experimental method:
[0036] 1. Biopanning procedure
[0037] 1. Blocking: 250 μL of 3% (w / v) BSA-TBS solution was added to each well to block the polystyrene microwell plate at 37° C. for 2 h.
[0038] 2. Incubation: After discarding the blocking solution and washing quickly, add 100 μL of the phage dodecapeptide library to the microwell plate and incubate at 37°C for 1 hour.
[0039] 3. Non-specific washing: the microwells were washed 5 to 15 times with 0.05% (v / v) Tween-TBS solution, and then washed 3 times with TBS.
[0040] 4. Specific elution: Discard the washing solution, add 150 μL of 100 mM HCl-Glycine (pH 2.2) solution for 10 min, and immediately add 9 μL of 2M Tris-HCl (pH 9.1) solution to neutralize the eluted phage.
[0041] 5. Amplification: Take 5 μL of this solution for phage titer determination and put the remaining solution into 25 mL of ER2738 bacterial solution in ...
Embodiment 2
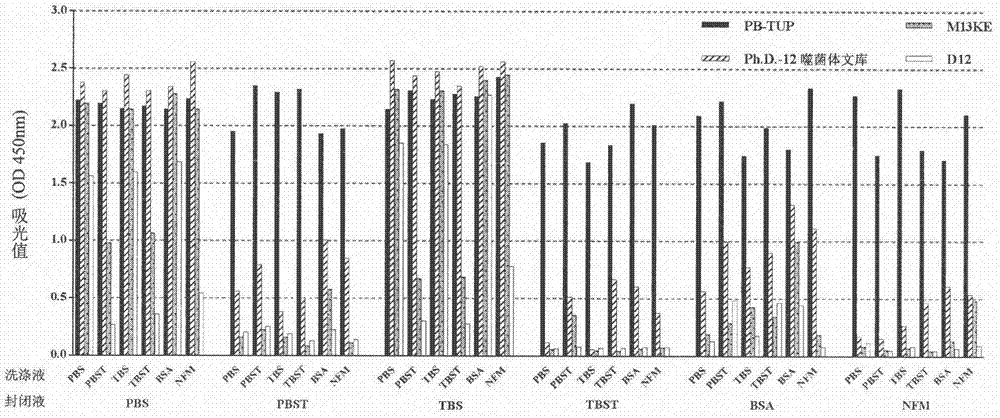
[0061] Comparison of the binding ability of each phage clone to polystyrene material under the conditions of different blocking solutions and washing solutions in the enzyme-linked immunosorbent assay
[0062] experimental method:
[0063] 1. Preparation of solutions: Six different solutions of PBS, 0.05% PBST, TBS, 0.05% TBST, 3% BSA-TBS and 3% NFM-TBS were prepared as blocking and washing solutions, respectively.
[0064] 2. Blocking: Use the above six solutions to block the polystyrene microplate at 37°C for 2 hours.
[0065] 3. Incubation of Phage: 10 10 (pfu / mL) PB-TUP phage clone, phage dodecapeptide library, wild-type M13KE phage clone and irrelevant peptide phage clone D12 were used as the experimental group and the control group, and were added to each polystyrene microwell treated with different blocking solutions. , and incubated overnight at 4°C.
[0066] 4. Washing: wash the above six solutions as different washing solutions for 5 times respectively
[0067] 5...
Embodiment 3
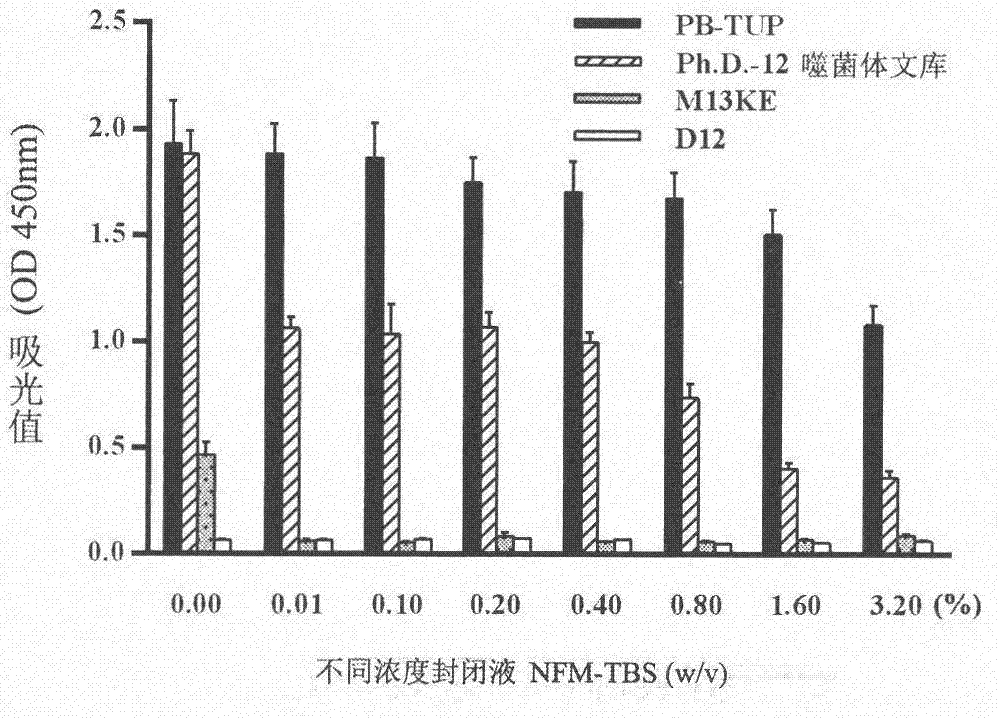
[0071] Eliminates the possibility of false positives due to phage adsorption to proteins in the blocking solution skim milk
[0072] experimental method:
[0073] 1. Blocking: NFM-TBS with different concentrations (0.00% to 3.20%) was added to each well and blocked at 37°C for 2 hours.
[0074] 2. Incubation of Phage: Bring the titer to 10 10 (pfu / mL) PB-TUP phage clone, phage dodecapeptide library, wild-type M13KE phage clone and irrelevant peptide phage clone D12 were used as the experimental group and the control group. Incubate overnight at 4 °C.
[0075] 3. Washing: Wash the microwells 5 times with 0.05% TBST solution.
[0076] 4. Incubation and detection of secondary antibody: add secondary antibody to incubate and carry out the subsequent steps of ELISA detection in Example 1.
[0077] Experimental results:
[0078] The result is as figure 2 As shown, NFM-TBS blocking solution inhibited all phage clones and polystyrene in a concentration-dependent manner. The bi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com