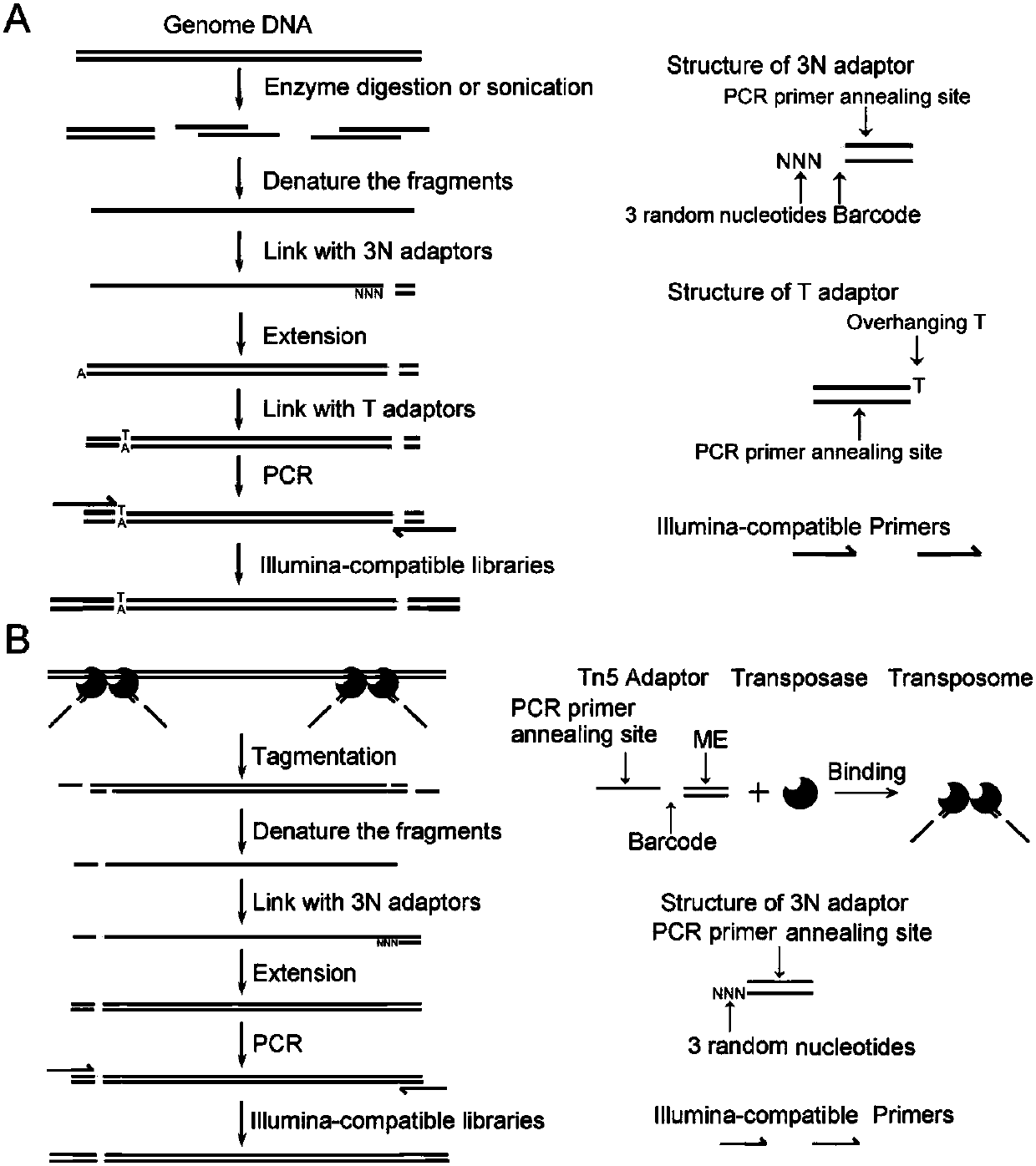
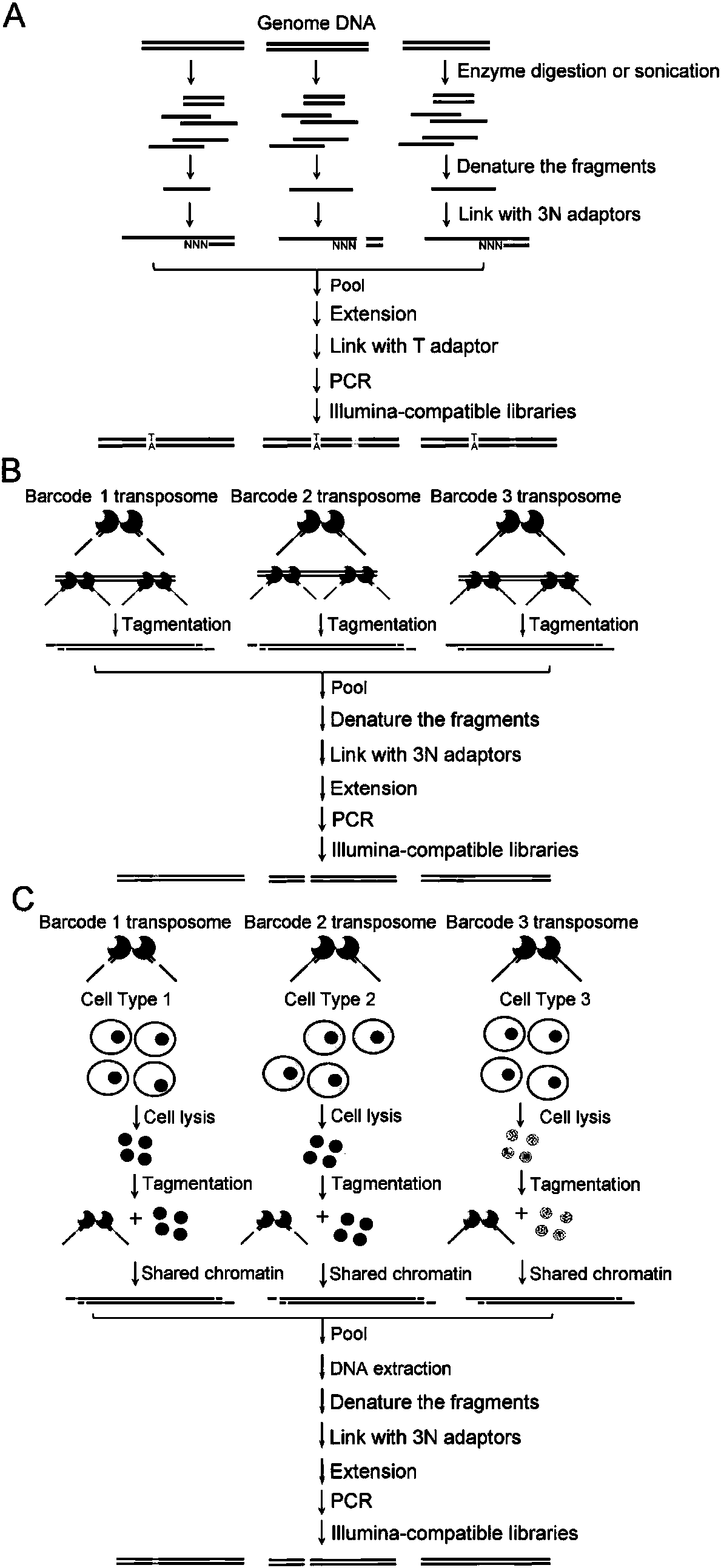
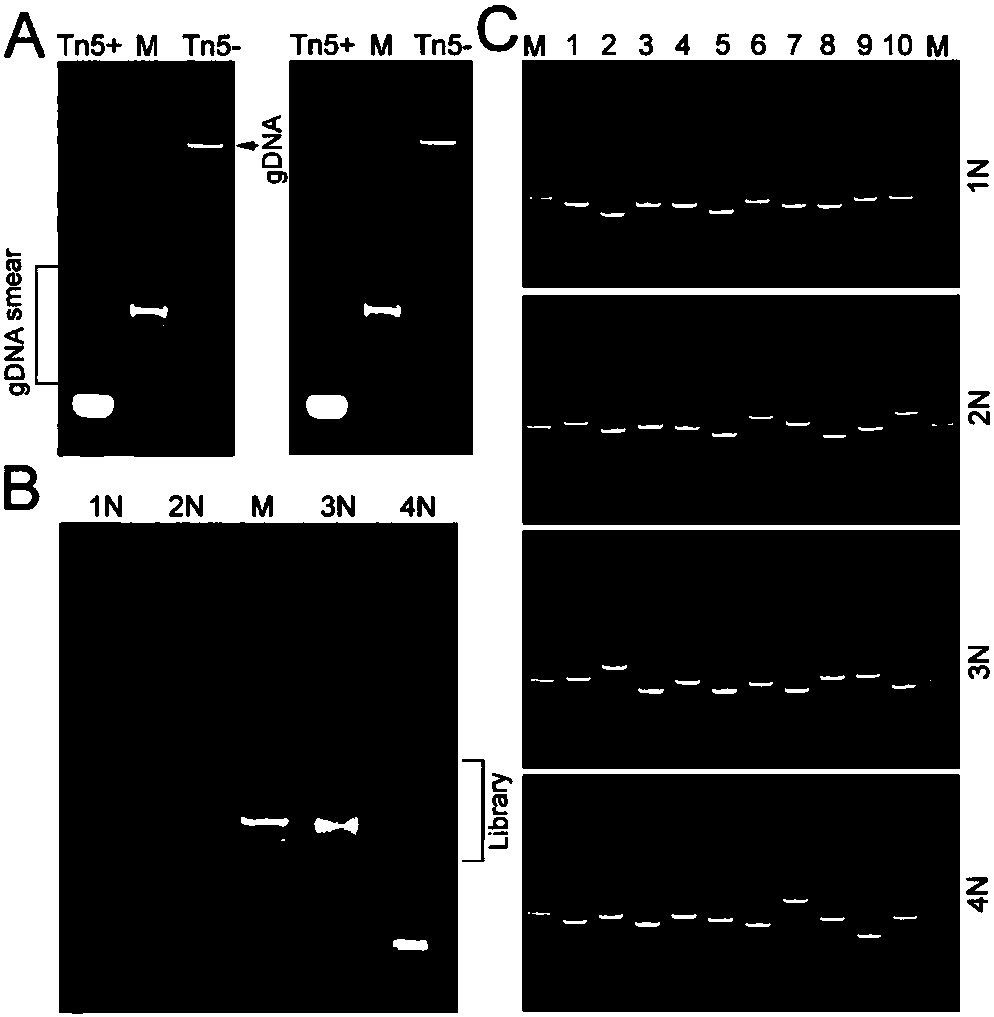
Construction method for next-generation sequencing library based on single-stranded connector and application thereof
A sequencing library and construction method technology, applied in the field of biomedicine, can solve problems such as loss of open area information, inability to detect DNA fragment sequences, etc., and achieve the effects of facilitating comparison and analysis, eliminating deviations, and simplifying library construction operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1 Based on the SALP of Tn5 transposome incisive reaction
[0061] experimental method:
[0062] Cell culture: HepG2 cells were cultured in DMEM medium. The medium contained 10% (v / v) fetal bovine serum, 100 units / mL penicillin and 100 μg / mL streptomycin. Cells at 37°C, 5% (v / v) CO 2 cultured in an incubator. Cells were obtained from the Cell Resource Center, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
[0063] Genomic DNA preparation: Genomic DNA (gDNA) of HepG2 cells was extracted by phenol-chloroform extraction.
[0064] Adapter preparation: All oligonucleotides were synthesized by Shanghai Sangon (as shown in SEQ ID NO.3-23 in Table 1). Preparation of Tn5 tag adapters (BTAs), tag and ME oligonucleotides with ddH 2 O was dissolved to 20 μM and mixed equimolarly to PCR tubes. Preparation of single-stranded linker (SSA), SSA-PN and SSA-PNre with ddH 2 O was dissolved to 100 μM and mixed equimolarly to PCR tubes. The above o...
Embodiment 2
[0074] Example 2 SALP-seq identification of chromatin opening state in different cell lines
[0075] experimental method:
[0076] Cell culture: HeLa, HepG2 and 293T were cultured in DMEM medium, and GM12878 cells were cultured in RPMI 1640 medium containing 10% (v / v) fetal bovine serum, 100 units / mL penicillin and 100 μg / mL streptomycin. Cells at 37°C, 5% (v / v) CO 2 cultured in an incubator. Cells were obtained from the Cell Resource Center, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
[0077] Adapter preparation: All oligonucleotides were synthesized by Shanghai Sangon (Table 1). Preparation of Tn5 tag adapters (BTAs), tag and ME oligonucleotides with ddH 2 O was dissolved to 20 μM and mixed equimolarly to PCR tubes. Preparation of single-stranded linker (SSA), SSA-PN and SSA-PNre with ddH 2 O was dissolved to 100 μM and mixed equimolarly to PCR tubes. The above oligonucleotide mixture was denatured at 95°C for 5 minutes, and then cooled ...
Embodiment 3
[0091] Example 3 SALP-seq identifies the open state of chromatin through different cell numbers
[0092] experimental method:
[0093] Cell culture: HepG2 was cultured in DMEM medium. The medium contained 10% (v / v) fetal bovine serum, 100 units / mL penicillin and 100 μg / mL streptomycin. Cells at 37°C, 5% (v / v) CO 2 cultured in an incubator.
[0094] Adapter preparation: All oligonucleotides were synthesized by Shanghai Sangon (Table 1). Preparation of Tn5 tag adapters (BTAs), tag and ME oligonucleotides with ddH 2 O was dissolved to 20 μM and mixed equimolarly to PCR tubes. Preparation of single-stranded linker (SSA), SSA-PN and SSA-PNre with ddH 2 O was dissolved to 100 μM and mixed equimolarly to PCR tubes. The above oligonucleotide mixture was denatured at 95°C for 5 minutes, and then cooled down to 25°C naturally.
[0095] Tn5 transposome preparation: According to the instructions of Robust Tn5 Transposase (Robust Tn5 Transposase, Robustnique Corporation Ltd.), mix ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com