Complete set reagent for detecting staphylococcus haemolyticus
A staphylococcus and reagent technology, applied in the field of complete sets of reagents for the detection of hemolytic staphylococcus, to achieve high sensitivity, good repeatability, amplification efficiency, and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Embodiment 1, the preparation of the complete set of reagents that detects staphylococcus hemolyticus
[0054] The complete set of reagents for detecting hemolytic staphylococcus consists of primer pairs and probe P for detecting or assisting the detection of hemolytic staphylococcus;
[0055] The primer pair is composed of single-stranded DNA named F and R respectively; F is the single-stranded DNA molecule shown in sequence 1 of the sequence listing; R is the single-stranded DNA molecule shown in sequence 2 of the sequence listing;
[0056] The sequence of the probe P is the sequence 3 in the sequence list, the 5' end of the probe is marked with a FAM fluorescent group, and the 3' end is marked with a non-fluorescence quenching group NFQ-MGB.
[0057] The first position of sequence 1-sequence 3 is the 5' terminal nucleotide of the corresponding sequence.
[0058] Each single-stranded DNA and probe of the complete set of reagents for detecting hemolytic staphylococcus a...
Embodiment 2
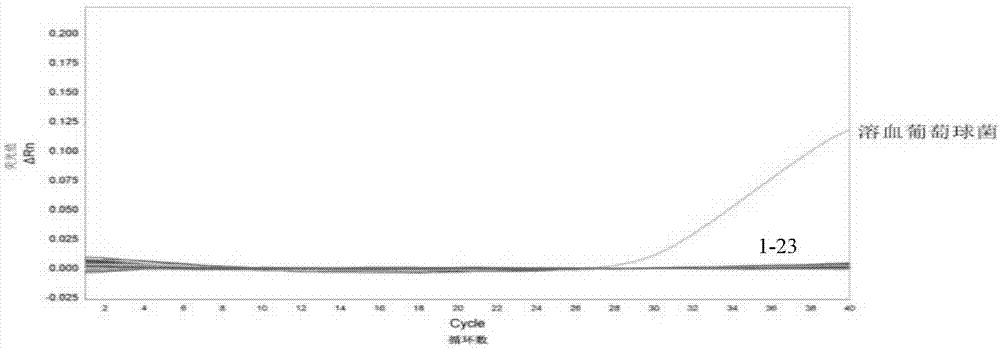
[0059] Embodiment 2, detect the specificity of the complete set of reagents of staphylococcus hemolyticus
[0060] 1. Strains to be tested
[0061] The strains used are the following 23 strains:
[0062] Ten species of Gram-positive bacteria (Streptococcus agalactiae, Staphylococcus hemolyticus (Shu Xiaoli; Wu Yidong; Shang Shiqiang, Establishment of a real-time fluorescence quantitative and typing method for bacterial 16SrRNA gene. Chinese Journal of Contemporary Pediatrics, 2008.10(6): p. 732-736.), Staphylococcus aureus (Li, L., et al., Phenol-soluble modulin alpha4 mediates Staphylococcus aureus-associated vascular leakage by stimulating heparin-binding protein release from neutrophils. Sci Rep, 2016.6: p.29373. ), Streptococcus pyogenes (Wang, J., et al., Identification and cluster analysis of Streptococcuspyogenes by MALDI-TOF mass spectrometry.PLoS One, 2012.7(11): p.e47152.), Streptococcus pneumoniae (Shu Xiaoli; Wu Yidong ; Shang Shiqiang, Establishment of a real-ti...
Embodiment 3
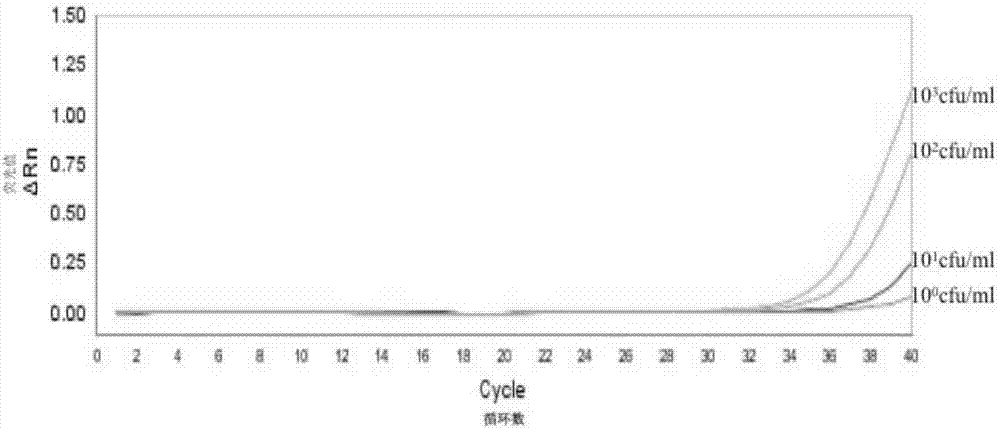
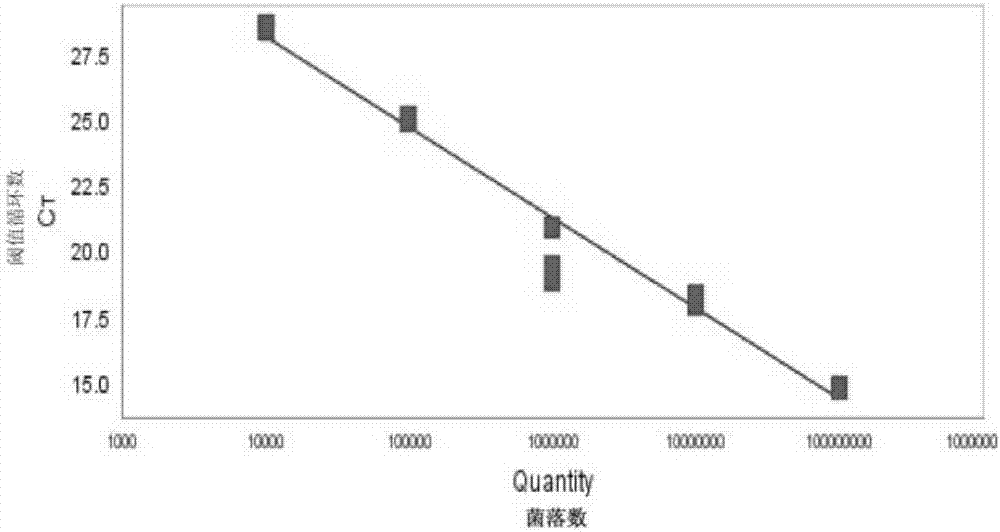
[0071] Embodiment 3, detect the sensitivity of the complete set of reagents of staphylococcus hemolyticus
[0072] After the hemolytic staphylococcus was quantified, a 10-fold serial dilution was performed to obtain a concentration of 10 3 cfu / mL, 10 2 cfu / mL, 10 1 cfu / mL and 10 0 For each cfu / mL strain suspension, 1 mL of each strain suspension was used to extract genomic DNA using the same method, and eluted with 80 μL elution buffer to collect the genome, and 5 μL was used as a template.
[0073] According to the reaction system and reaction procedure of Example 2, the sensitivity of the kit of reagents in Example 1 was tested using the above genome solutions as templates, and three replicates were set for each concentration of genome solutions.
[0074] result( figure 2 ) shows that 10 3 cfu / mL, 10 2 cfu / mL, 10 1 cfu / mL and 10 0 There are amplification curves in the reaction system of cfu / mL, show that, the complete set of reagents of embodiment 1 can detect 10 0...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com