Micro-RNA-21 ultra-sensitive detection method based on double-enzyme signal cascade amplification
An RNA-21 and cascade amplification technology, which is applied in the ultra-sensitive detection field of microRNA-21, can solve the problems that the stability of the probe is not optimal and cannot be commercialized, and achieves great application value and high sensitivity. and selective, easy-to-use effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] (1) Mix carboxylated polystyrene microspheres and 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride aqueous solution in phosphate buffer at a ratio of 1:4, and react at room temperature After 15 to 30 minutes, add DNA single strand (one end is aminated and the other end is carboxylated), and react overnight at room temperature; react by coupling carboxylamino group and centrifuge for purification to obtain a DNA-labeled probe.
[0046] (2) The DNA probe and horseradish peroxidase are then activated and coupled at a molar ratio of 1:100, and reacted for 2 to 3 hours at room temperature; Enzyme-labeled probes.
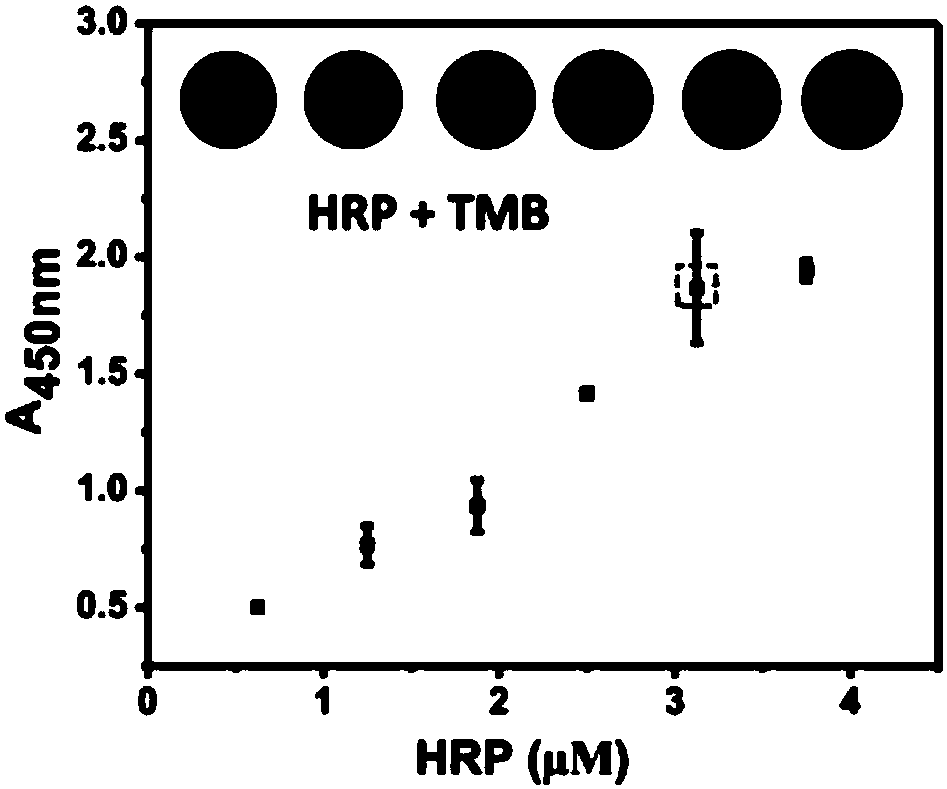
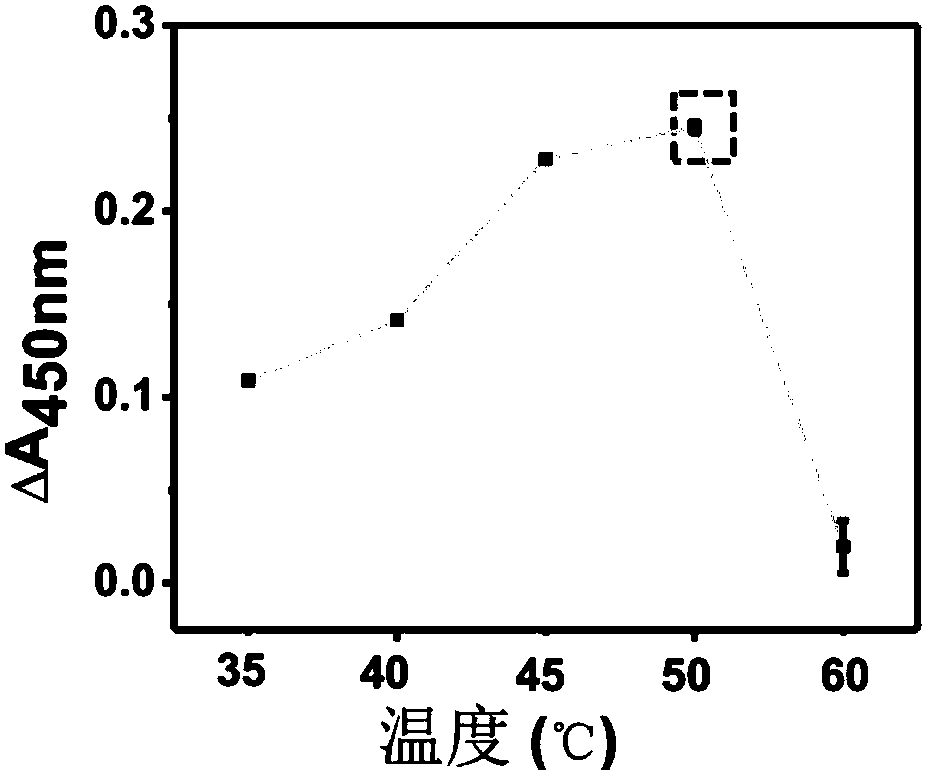
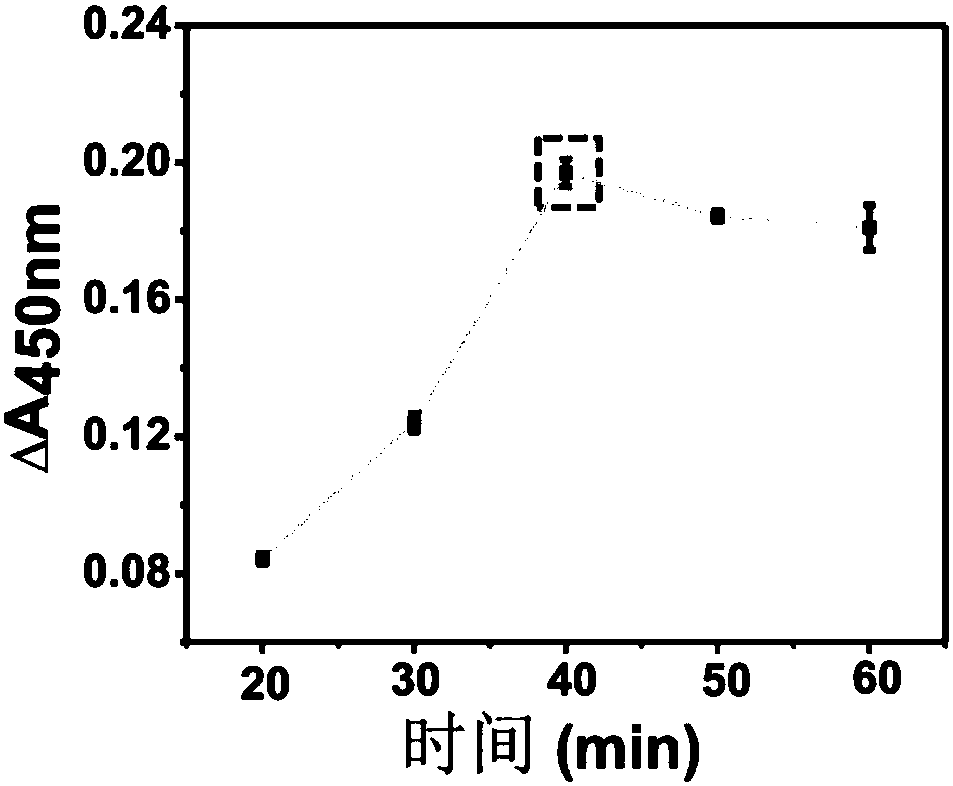
[0047] (3) Mix equal volumes of the above-mentioned probe and TMB one-component chromogenic solution, and react in the dark for 10-15 minutes at room temperature. An equal volume of hydrochloric acid was added to terminate the reaction, and the absorbance of the solution at 450 nm was detected with a microplate reader. According to the change of absorb...
Embodiment 2
[0050] (1) Mix carboxylated polystyrene microspheres and 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride aqueous solution in phosphate buffer at a ratio of 1:4, and react at room temperature After 15 to 30 minutes, add DNA single strand (one end is aminated and the other end is carboxylated), and react overnight at room temperature; react by coupling carboxylamino group and centrifuge for purification to obtain a DNA-labeled probe.
[0051] (2) Then the DNA probe and horseradish peroxidase are activated and coupled according to the molar ratio of 1:200, and reacted at room temperature for 2 to 3 hours; Enzyme-labeled probes.
[0052] (3) Mix equal volumes of the above-mentioned probe and TMB one-component chromogenic solution, and react in the dark for 10-15 minutes at room temperature. An equal volume of hydrochloric acid was added to terminate the reaction, and the absorbance of the solution at 450 nm was detected with a microplate reader. According to the chan...
Embodiment 3
[0055] (1) Mix carboxylated polystyrene microspheres and 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride aqueous solution in phosphate buffer at a ratio of 1:4, and react at room temperature After 15 to 30 minutes, add DNA single strand (one end is aminated and the other end is carboxylated), and react overnight at room temperature; react by coupling carboxylamino group and centrifuge for purification to obtain a DNA-labeled probe.
[0056] (2) Then the DNA probe and horseradish peroxidase are activated and coupled according to the molar ratio of 1:300, and reacted at room temperature for 2 to 3 hours; Enzyme-labeled probes.
[0057] (3) Mix equal volumes of the above-mentioned probe and TMB one-component chromogenic solution, and react in the dark for 10-15 minutes at room temperature. An equal volume of hydrochloric acid was added to terminate the reaction, and the absorbance of the solution at 450 nm was detected with a microplate reader. According to the chan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com