Method for improving mass spectrometry detection microorganism secondary crystal, and product
A mass spectrometry detection and microorganism technology, applied in the field of mass spectrometry detection, can solve the problems of poor crystal morphology, inability to save time and cost, and complex preparation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment one, the preparation of matrix solution
[0063] The main component of the matrix solution is 3-hydroxy-2-pyridinecarboxylic acid, and a certain proportion of acetonitrile is added to accelerate the volatilization of the primary crystals of the matrix and quickly form uniform and intact primary crystals.
[0064] The preparation steps of matrix solution are as follows:
[0065] A, using deionized water and high performance liquid chromatography grade acetonitrile according to V deionized water: V acetonitrile = 1:1-0.5:1 volume ratio mixed to obtain mixed solution I;
[0066] B. Weigh 3-hydroxy-2-pyridinecarboxylic acid and place it in a centrifuge tube, then add mixed solution I to dissolve, wherein the weight mg of 3-hydroxy-2-pyridinecarboxylic acid and the mixed solution volume mL ratio is 1-10:100 ;
[0067] C. Shake at 2000-3000rpm for 3-10min, centrifuge at 8000-12000rpm for 3-10min to obtain 3-hydroxy-2-pyridinecarboxylic acid solution;
[0068] D....
Embodiment 2
[0076] Example 2, primary crystallization form of biological samples
[0077] (1) Position the hole in the hydrophilic area, point 0.5-1 μL of matrix solution, and form a crystal after natural evaporation;
[0078] (2) In the central calibration hole, place 0.5-1 μL of matrix solution, and form a primary crystal after natural evaporation;
[0079] (3) To verify the calibration hole, point 0.5-1 μL of matrix solution, and form a crystal after natural evaporation;
[0080] Such as figure 1 The left picture is the crystallization map of the positioning hole in the hydrophilic region, and the right picture is the crystallization map of the center correction hole. The shape of the two crystals is regular and round, the surface is round like jade, the texture is regular and uniform, and the crystals grow meticulously in all directions, which is an ideal protein crystal form.
[0081] It should be pointed out that Embodiments 1 and 2 should be carried out in the operating room wit...
Embodiment 3
[0082] Example 3. Determining the optimal ratio of matrix to sample solution through secondary crystallization
[0083] According to the principle of matrix-assisted laser desorption ionization, a standard biological sample (Escherichia coli 8739) was selected, and 0.5, 0.75, and 1 μL of matrix solution were placed in the middle of three wells, and a crystallization was formed after natural evaporation;
[0084] Then, on the primary crystallization surface, evenly point 0.5 μL standard sample, and form secondary crystallization after natural evaporation;
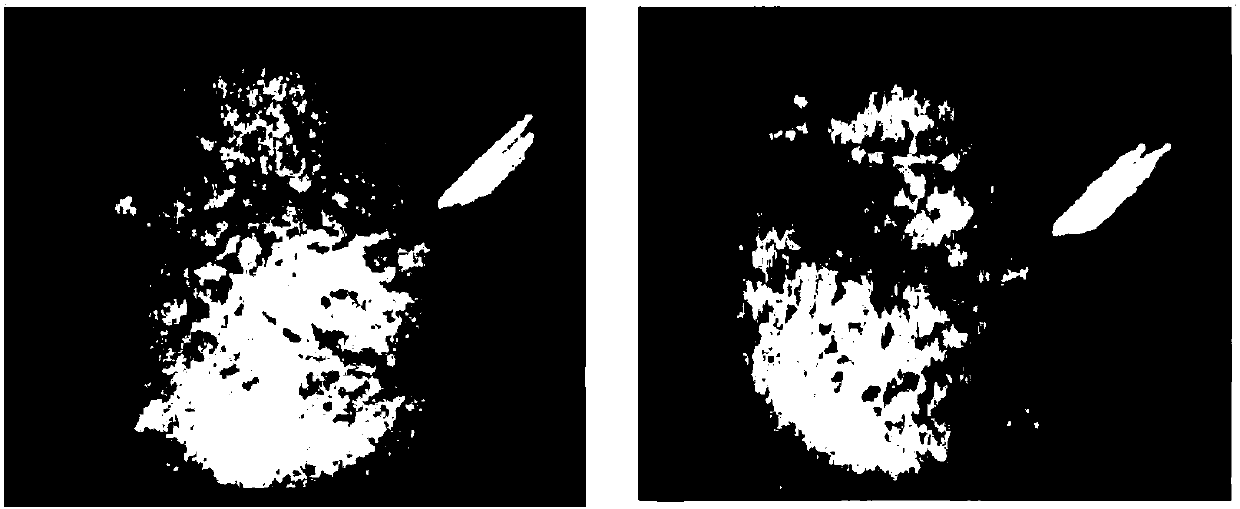
[0085] The comparison of secondary crystallization is as figure 2 shown. Among them, the crystals of 0.5 μL and 1 μL matrix solutions were irregular, with uneven thickness, hollow in the middle and thick around. Therefore, when the laser bombards the sample, the sample peak accuracy is poor, the noise is high, and the baseline is high.
[0086] The 0.75 μL matrix solution is regular, uniform in thickness, fine in crystal...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com