DNA methylation detection kit and application thereof
A detection kit and methylation technology, applied in the field of molecular biology, can solve problems such as low recovery rate and loss of genomic DNA fragments, achieve high recovery efficiency, good integrity, and save a lot of time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Preparation and preparation of the kit:
[0052] CT conversion solution: 2.5mol / L Na 2 S 2 O 5 , NaOH adjusts the pH of the solution to 5.0
[0053] Antioxidant: 10mM C 6 H 4 (OH) 2 (Quinol)
[0054] Binding solution: 4mol / L Gu-HCl (guanidine hydrochloride), 1mol / L KAC, adjust the pH of the solution to 4.5
[0055] Washing solution: 70% ethanol
[0056] Desulfonation solution: 100mM NaOH, 70% ethanol
[0057] Eluent: 10mM Tris, 1mM EDTA, pH8.0
[0058] Magnetic bead suspension: carboxyl magnetic beads with a particle size of 200nm
[0059] The CT conversion solution, antioxidant, binding solution, desulfonation solution and eluent solvent are all double distilled water.
[0060] After the above reagents are configured separately, the kit is assembled.
[0061] The following detailed steps are applicable to the operation of the kit of the present invention:
[0062] Step 1. Add 0.2-2μg human gDNA to the PCR tube. If the volume of the DNA sample is less than 20ul, add ddH 2 O to 20μl.
[...
example
[0081] There is no significant difference in the effect of using the kit within the above range. For parallel comparison, the following examples uniformly adopt the following steps:
[0082] Step 1. Add 0.5μg human gDNA to the PCR tube and add ddH 2 O to 20μl.
[0083] Step 2. Add 208 μl of CT conversion solution and 12 μl of antioxidant to the above tube, mix thoroughly and gently, and react according to the following procedure: 98°C for 10 minutes; 98°C for 30s and 60°C for 150 minutes.
[0084] Step 3. After the reaction is completed, transfer the above reaction solution to a 1.5 ml EP tube.
[0085] Step 4. Add 240 μl binding solution and 30 μl magnetic bead suspension to the above reaction solution, shake and mix thoroughly, combine for 3 minutes at room temperature, and mix occasionally.
[0086] Step 5. Place the above EP tube on the magnetic separation rack for magnetic separation, aspirate the supernatant, save the magnetic beads, and take out the centrifuge tube from the magn...
Embodiment 1-1
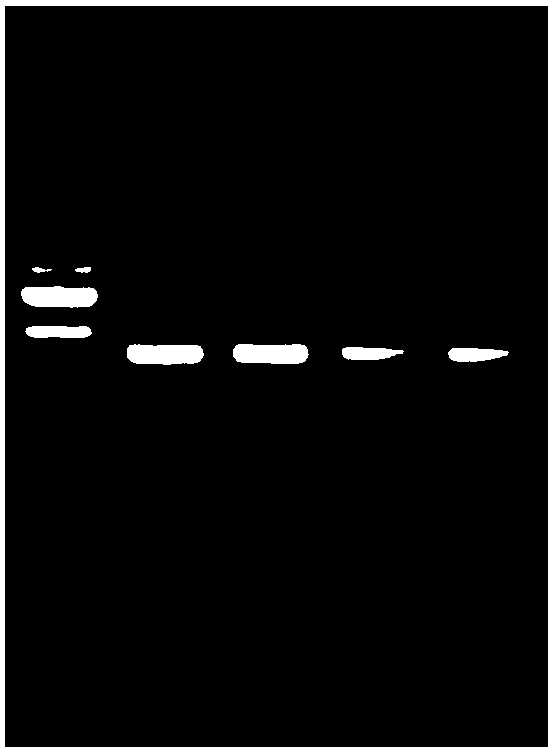
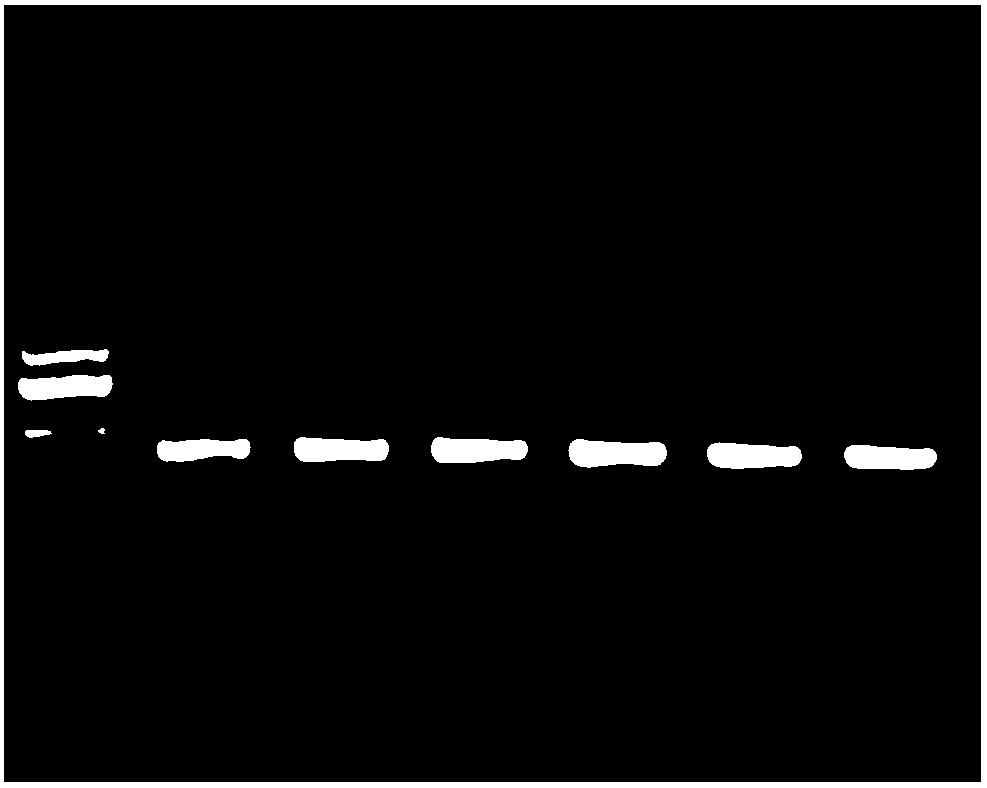
[0098] In this example, the kit of the present invention is used to detect whether a segment of the promoter region of the human phosphatase and tensin homolog (PTEN) gene is methylated. PTEN is a tumor suppressor gene that can negatively regulate the growth of tumor cells. Human genomic DNA is extracted from cultured gastric cancer cells using the Magnetic Bead Method Animal Tissue Genomic Extraction Kit (B518721).
[0099] In order to confirm the reliability of the test results of the kit of the present invention, a commercially available similar product was used as a control kit to simultaneously process 500 ng of human cell genomic DNA.
[0100] experiment procedure:
[0101] BSP experiment:
[0102] Primer design: Take the sequence of the promoter region of the PTEN gene as the target sequence, use ABI's Methyl primerExpress software to analyze and find the CPG island in the promoter region, and then design a pair of BSP primers. The primer sequence is shown below.
[0103] BSP-F...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
| conversion efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com