Method for determining composition of disulfide bond of recombinant human granulocyte colony stimulating factor
A colony-stimulating factor and disulfide bond technology, applied in the field of proteomics, can solve problems such as poor repeatability, high requirements for experimental equipment, and complicated processing procedures, and achieve high vigor, low data analysis difficulty, and simple operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
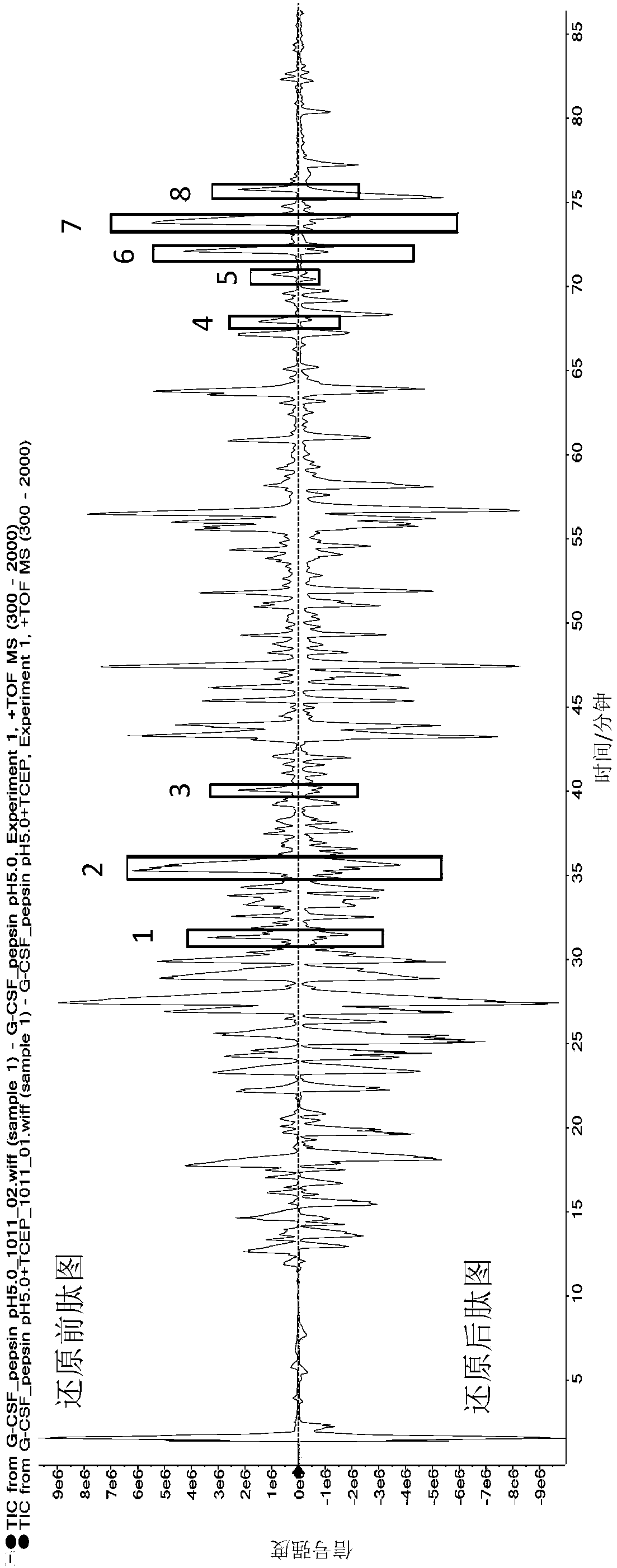
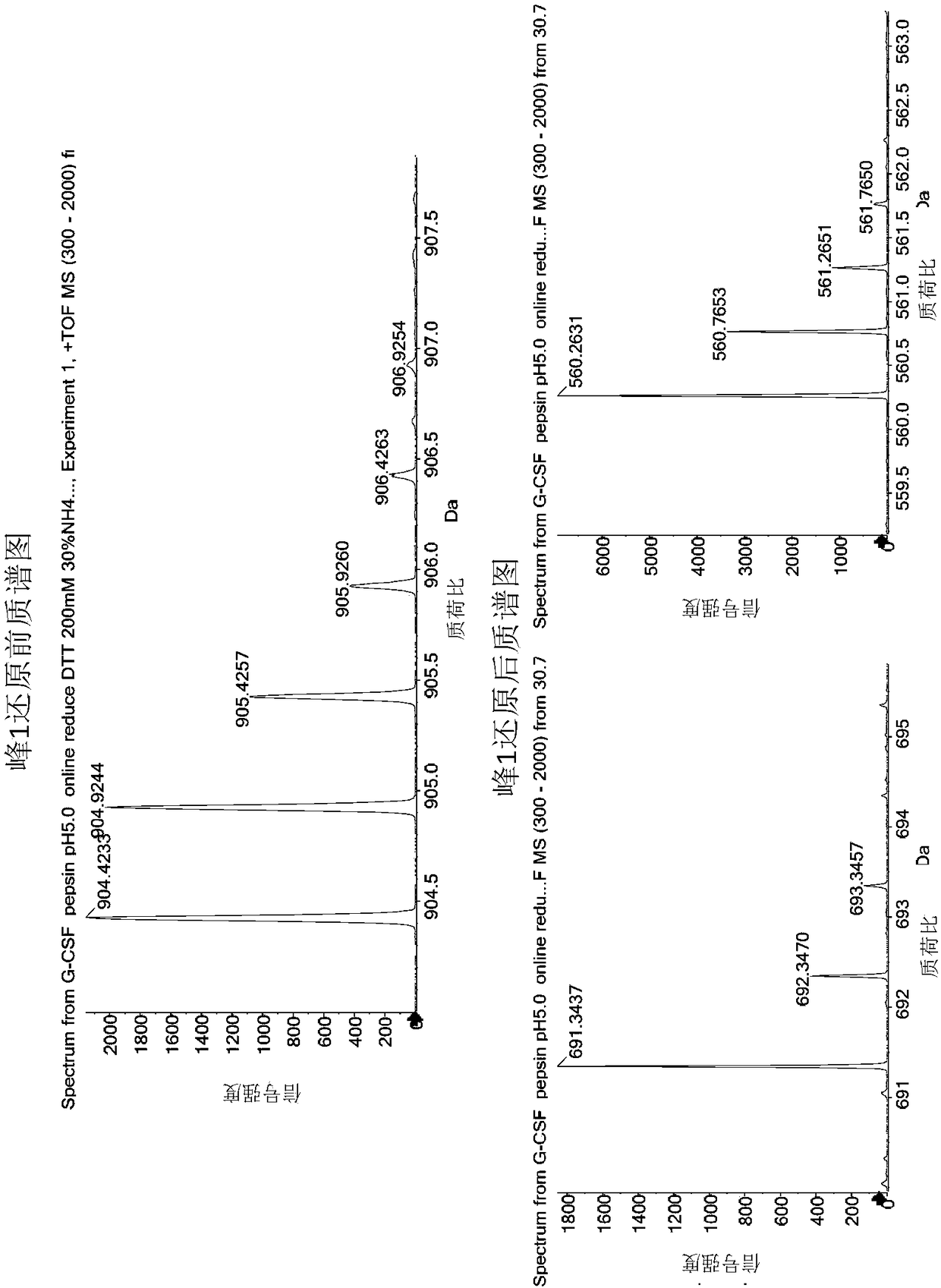
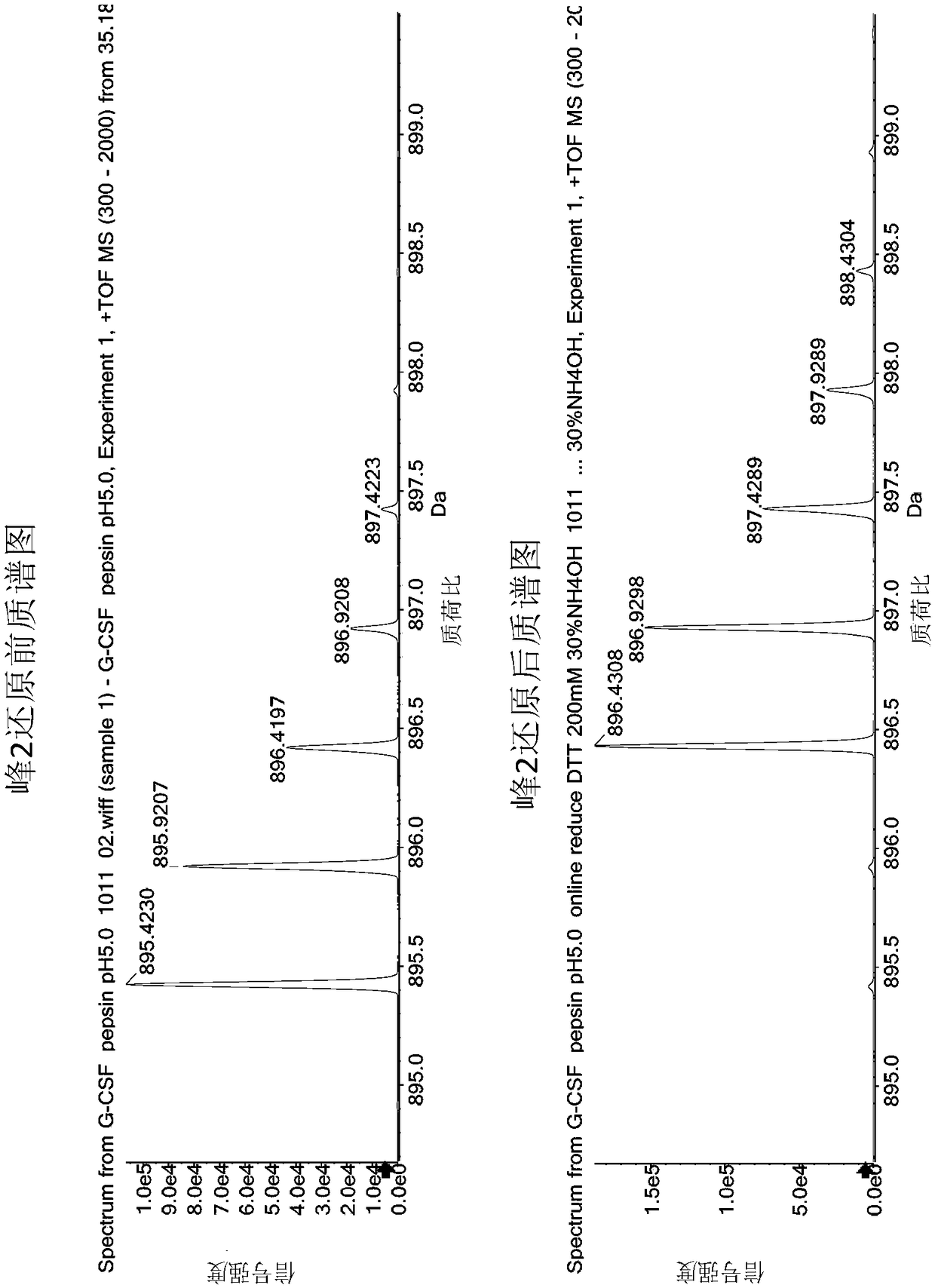
[0047] 1. Pepsin hydrolysis of rhG-CSF samples
[0048] Pepsin (purchased from Sigma, source: pig gastric mucosa) was prepared into a 1 mg / ml solution with ultrapure water, and each 100 μl tube was frozen and stored for future use. Prepare 50 mM citric acid solution, adjust to pH 5.0 with 0.1 mol / L sodium hydroxide solution, and filter with 0.45 μm microporous membrane. Use this solution to dilute the rhG-CSF stock solution to 1 mg / ml, add 1 mg / ml pepsin according to the volume ratio of 40:1, vortex and mix three times, each time for 5 seconds, and then place it in a 37°C water bath for 16 hours after mixing. Take 100 μL of the above enzymatic hydrolysis product and place it in a liquid chromatography sampling bottle, which is the sample before reduction. Take out 100 μL of the enzymatic hydrolysis product and put it in an EP tube, add 1 μL of TCEP solution (concentration: 1mol / L, prepared with ultrapure water), vortex and mix three times, each time for 5 seconds, mix well an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com