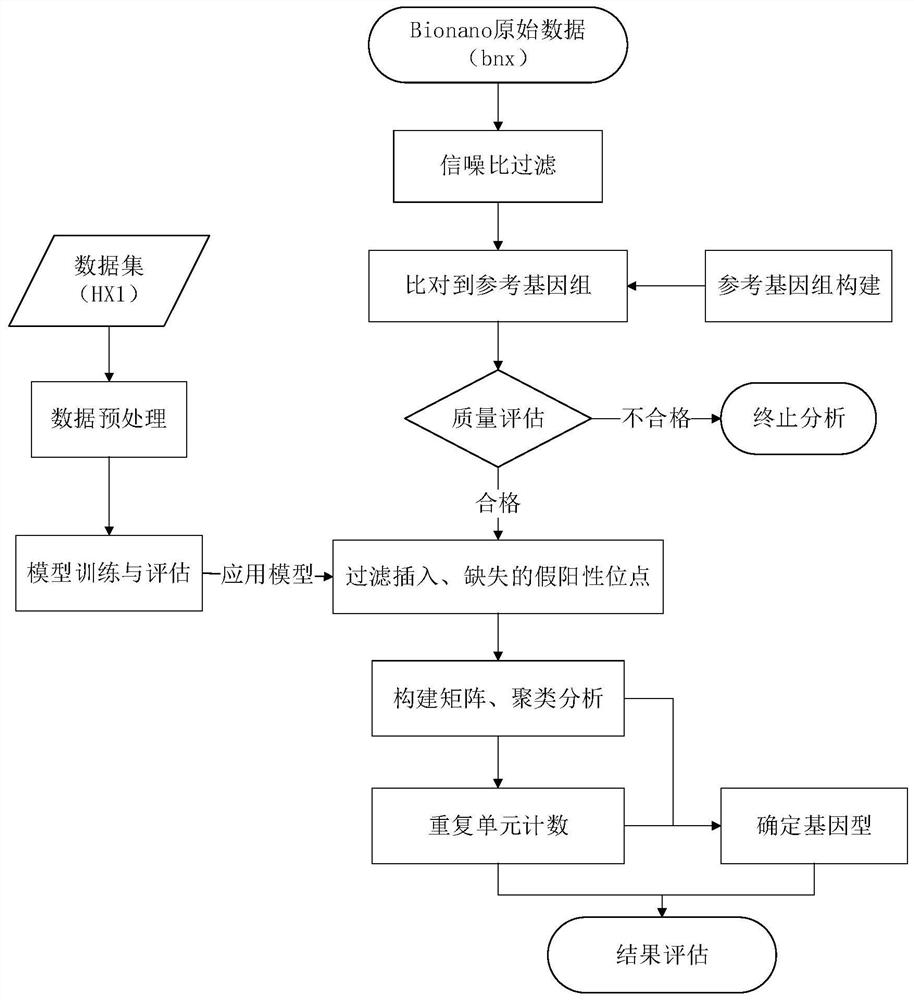
A method for detecting long tandem repeats based on the bionano platform
A technology of tandem repeat sequences and platforms, applied in genomics, instrumentation, proteomics, etc., can solve the problems of assembly errors, consumption of computing resources, long computing time, etc., to achieve fast running speed, consume less memory resources, and improve accuracy. sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
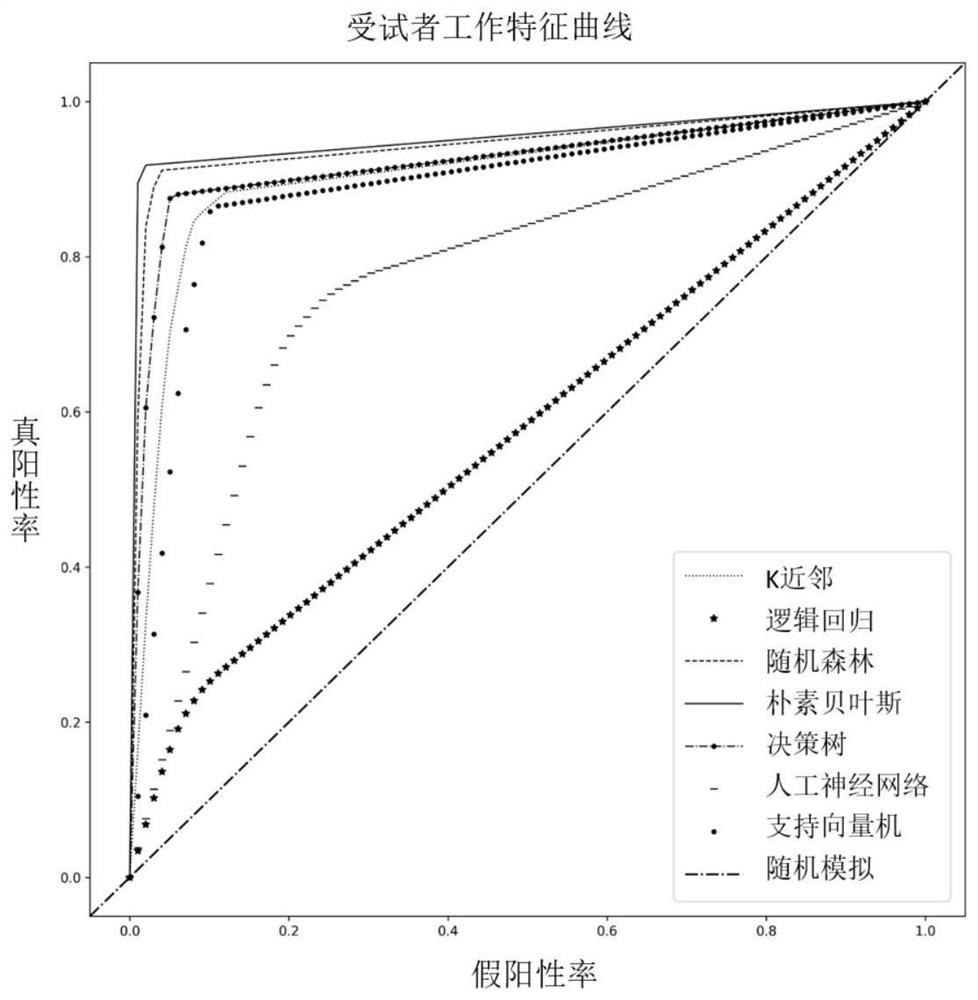
[0065] Embodiment 1. Building a machine learning model
[0066] a. Dataset
[0067] Using HX1 data (see Shi, L., Guo, Y., Dong, C., Huddleston, J., Yang, H., Han, X. & Lintner, K.E. (2016). Long-read sequencing and de novo assembly of a Chinese genome.Nature communication.) constructed the Chinese reference genome and its Bionano optical map data, and compared the Bionano data to the HX1 reference genome, and the sites that could be compared to the reference genome were defined as true positive sites. The detected sites were defined as false positive sites. Accordingly, we randomly selected 1000 true positive sites and 1000 false positive sites as data sets for the two cases.
[0068] b. Feature selection
[0069] According to the data characteristics of Bionano, the intensity (Intensity), signal-to-noise ratio (SNR), and coverage of the site are weighted according to the confidence (Confidence) of reads compared to the reference genome (shown in formula 1-4). At the same ...
Embodiment 2
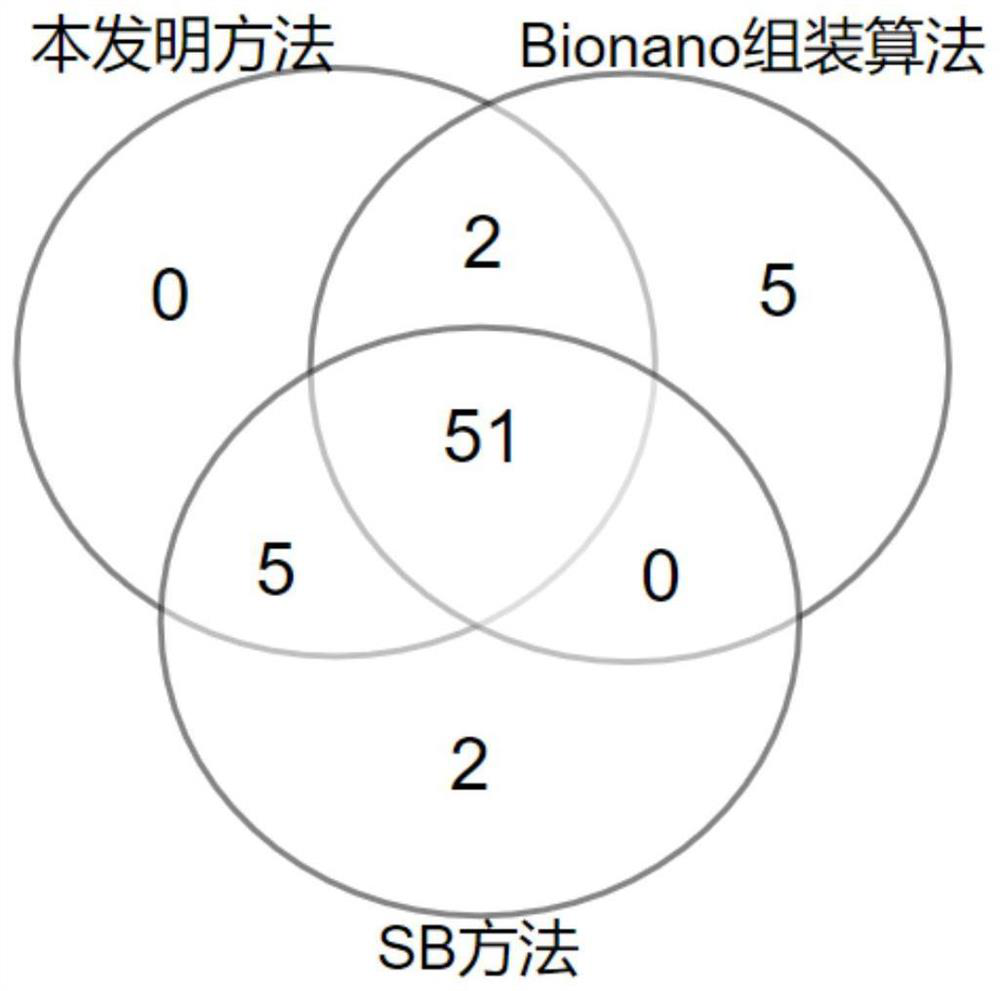
[0102] Example 2. Long tandem repeat sequence detection
[0103] 1. Experimental method
[0104] Lysis of human red blood cells (1hour)
[0105] Quantitative white blood cell count (5min)
[0106] Embedding of white blood cells (~1hour)
[0107] Digestion with proteinase K
[0108] Wash gel to immobilize DNA
[0109] DNA recovery
[0110] DNA dialysis and homogenization
[0111] Quantification of DNA concentration (10μl, 2 hours and 30 minutes)
[0112]Digest DNA with BssSI enzyme (10 μl, 2 hours and 30 minutes)
[0113] Labeling (15 μl, 1 hr 15 min)
[0114] Fix (20 μl, 45 minutes)
[0115] Staining treatment (60 μl, 16 hours / overnight)
[0116] BionanoSaphyr system for quantitative processing
[0117] Experiment details reference:
[0118] https: / / bionanogenomics.com / wp-content / uploads / 2017 / 03 / 30033-Rev-C-Bionano-Prep-Blood-DNA-Isolation-Protocol.pdf;
[0119] https: / / bionanogenomics.com / wp-content / uploads / 2017 / 07 / 30024-Rev-J-Bionano-Prep-Labeling-NLRS-Protocol....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com