Specific DNA aptamer for CD4 positive cell and chimera of specific DNA aptamer
A nucleic acid aptamer and positive cell technology, applied in the direction of recombinant DNA technology, DNA / RNA fragments, drug combination, etc., can solve the problem of high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
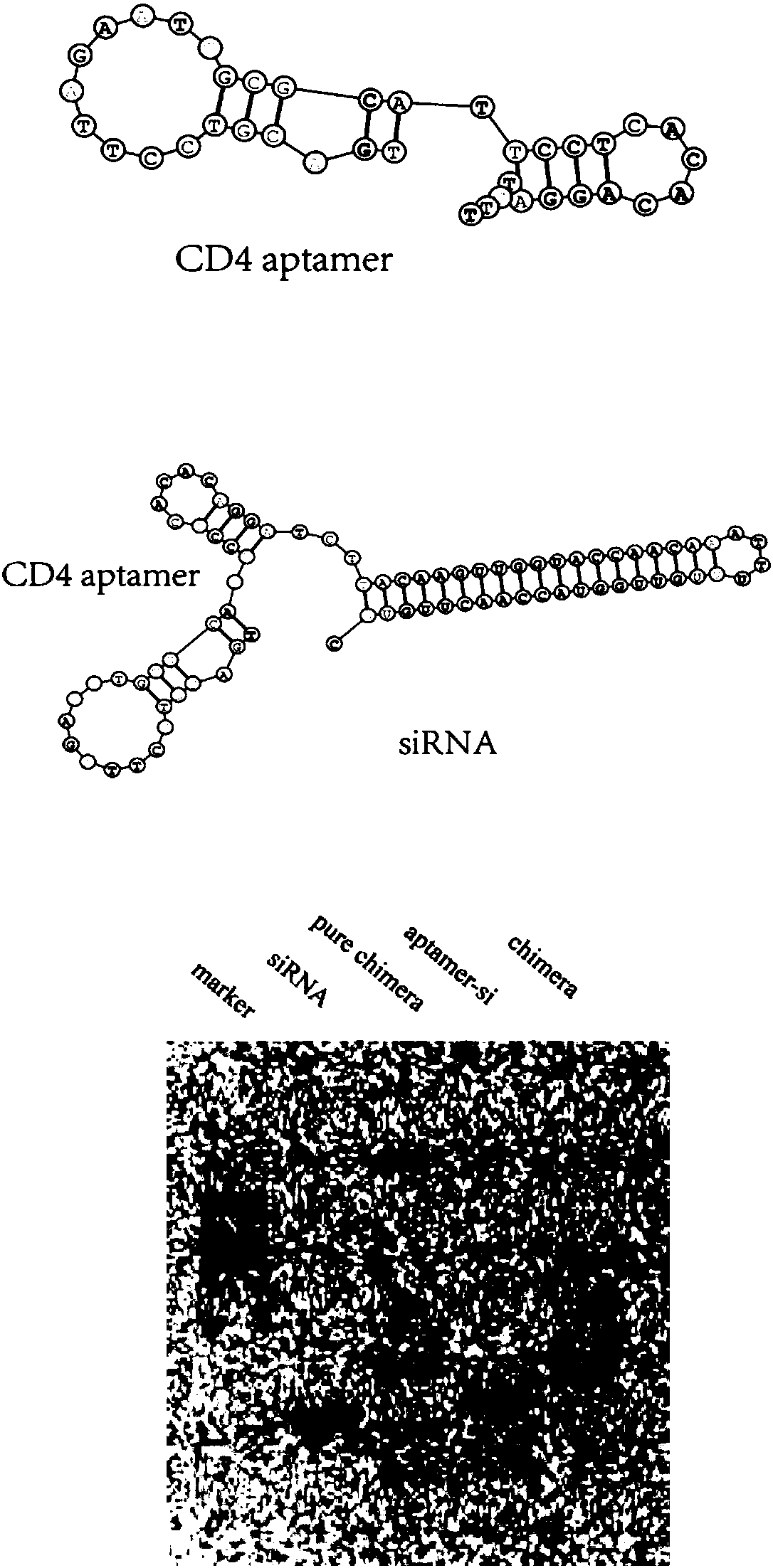
[0067] Example 1. Construction and verification of CD4 nucleic acid aptamer aptamer and CCL18 siRNA-connected chimera chimera
[0068] 1.1 CD4 DNA aptamer, aptamer-linked siRNA sense strand intermediate and siRNA antisense strand were synthesized and provided by the company (TAKARA, Gemma). Mix the same concentration of intermediates and siRNA antisense strands, add annealing buffer, slowly anneal to 25°C in a 90°C water bath, and store in minus 80°C.
[0069] 1.2 Add the same concentration of CD4 nucleic acid aptamer, intermediate, siRNA antisense strand, and chimera to the loading buffer, and electrophoresis at 150V for 10 minutes in 8% non-denaturing PAGE gel, and observe the position of the chimera constructed in the electrophoresis.
[0070] The secondary structure of the full-length aptamer is predicted in the secondary structure prediction software, and it can be found that the CD4 aptamer has two stem-loop structures such as figure 1 (Top), the morphology of the chime...
Embodiment 2
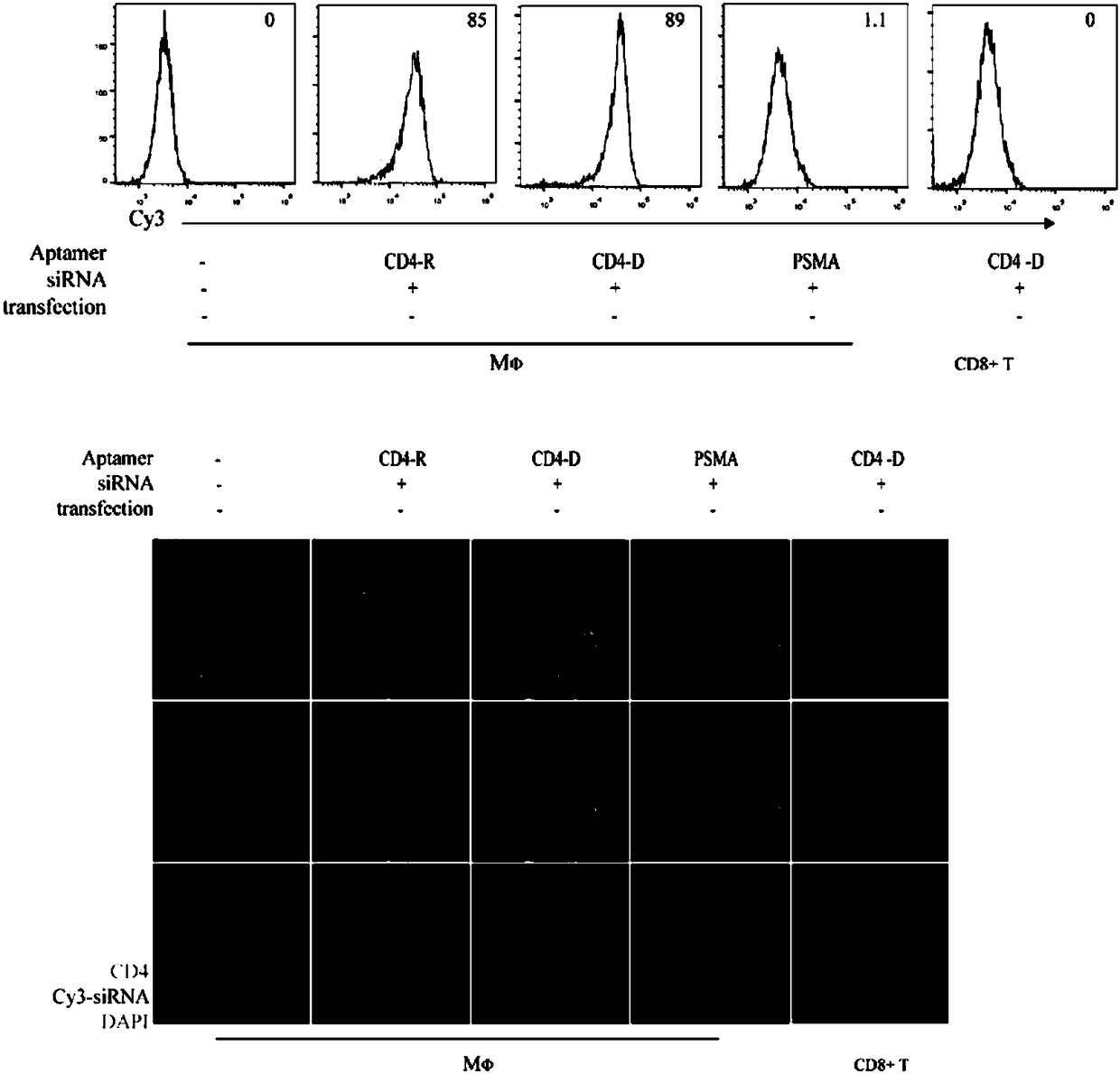
[0071] Example 2. Verification of uptake rate of chimeras
[0072]2.1 In the TAMs, add the CD4 nucleic acid aptamer labeled with the cy3 fluorescent group at a final concentration of 10 nM. Treat TAMs with an equal amount of PSMA nucleic acid aptamer (also labeled as cy3) as control 1, treat MDA-MB-231 cells with CD4 nucleic acid aptamer as control 2, and liposome transfect fluorescently labeled siRNA double-strand transfected TAMs served as control III.
[0073] 2.2 Collect the treated cells after 24 hours of treatment together, wash off the excess treatment reagent with PBS once, centrifuge at 300g for 5 minutes, discard the supernatant, add 200 microliters of PBS to resuspend, and analyze the uptake of aptamer or siRNA by the cells with a flow cytometer Case.
[0074] 2.3 Immunofluorescence detection: Before the above treatment, plant TAMs or MDA-MB-231 cells on a small glass slide, and perform the above treatment after the cells adhere to the wall. After 24 hours, the s...
Embodiment 3
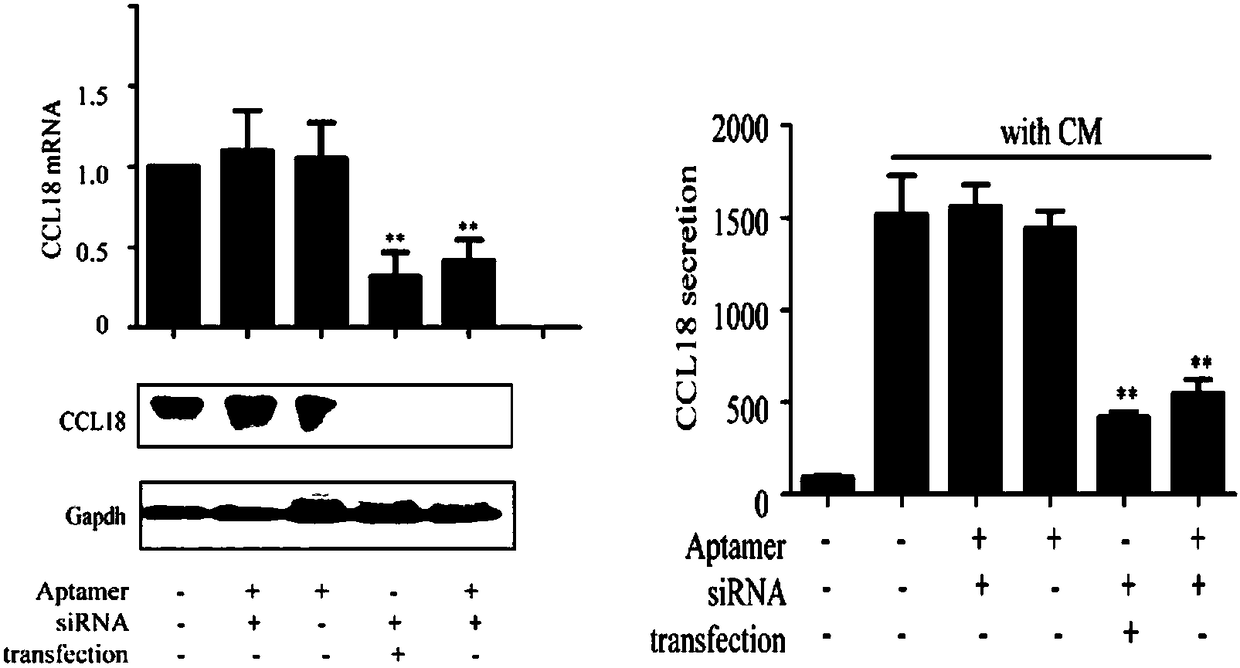
[0084] Example 3. The constructed chimera inhibits the synthesis and secretion of ccl18
[0085] 3.1 In TAMs, add chimeras at a final concentration of 20 nM, CD4 aptamer as a blank control, chimeras linked to GFP protein siRNA as a negative control, and liposome transfection as a positive control. After 24-36 hours of culture, add 1 ml of trizol to each well to lyse the cells and collect mRNA; after 48-60 hours of culture, collect cell protein and cell supernatant.
[0086] 3.2 RT-QPCR
[0087] 3.2.1 Add 200 microliters of chloroform to the cell mixture containing trizol, shake it vigorously and let it stand for 10 minutes, then centrifuge at 12,000 rpm at 4°C for 15 minutes; after centrifugation, absorb about 400 microliters of the upper transparent liquid phase and add 1 milliliter of isopropyl Alcohol, mix gently and let stand for 5 minutes, centrifuge at 12,000rpm at 4°C for 10 minutes; discard the supernatant after centrifugation, keep the bottom precipitate, add 1 ml of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com