PSR (Polymerase Spiral Reaction) detection primer for escherichia coli O157:H7, kit and detection method thereof
A detection kit, O157 technology, applied in the biological field, can solve the problems of high reagent price, complex primer design, high false positive rate, etc., and achieve the effect of low detection cost, short time-consuming, simple and fast operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1 detects the microbial method of E.coliO157:H7 based on polymerase helical reaction isothermal amplification technology
[0044] 1. The method for detecting pathogenic microorganisms based on polymerase helical isothermal amplification technology. In this embodiment, E.coliO157:H7 is used as an example, and the reagents used are as follows:
[0045] a. Detection primers Ft aqueous solution and Bt aqueous solution, accelerated primer IF and IB aqueous solution with a concentration of 50 μM each, the primer sequences are as follows (5'-3'):
[0046] Detection primer Ft: TTGGCATCGTGTGGACAGGGTAGGACCGCAGAGGAAAGA (SEQ ID NO.1);
[0047] Detection primer Bt: TGGGACAGGTGTGCTACGGTTTCCACGCCAACCAAGATC (SEQ ID NO.2);
[0048] Acceleration primer IF: GTTTCGATGAGTTTATCTGC (SEQ ID NO.3);
[0049] Acceleration primer IB: TCAAAAGCACCCTATAGCTG (SEQ ID NO.4);
[0050] b.2×Reaction stock solution: Tris-HCl with concentration of 40.0mM, ammonium sulfate of 20.0mM, potassium c...
Embodiment 2
[0060] Example 2 Polymerase helical reaction detection E.coli O157:H7 specificity test
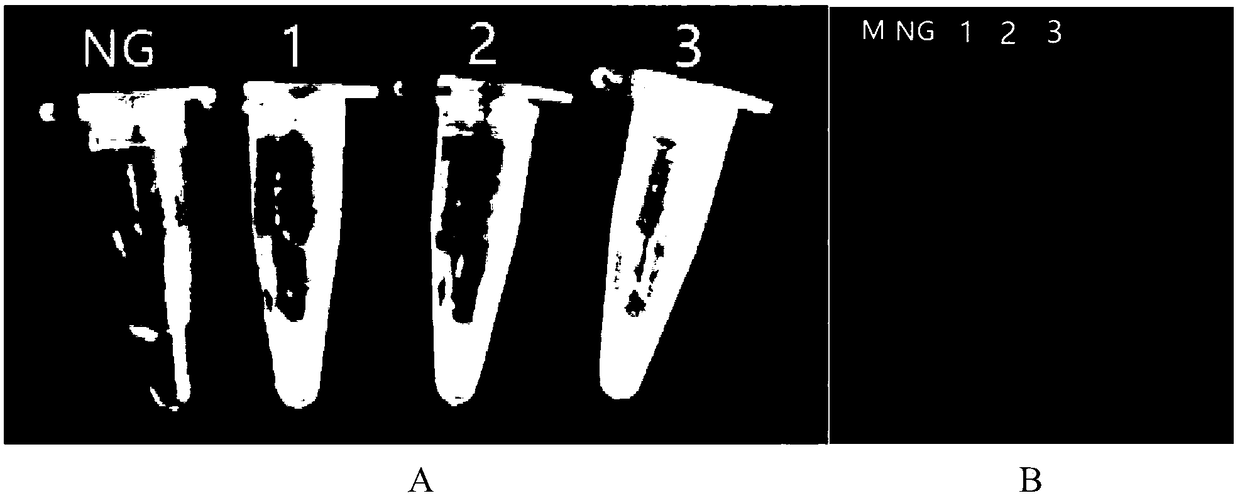
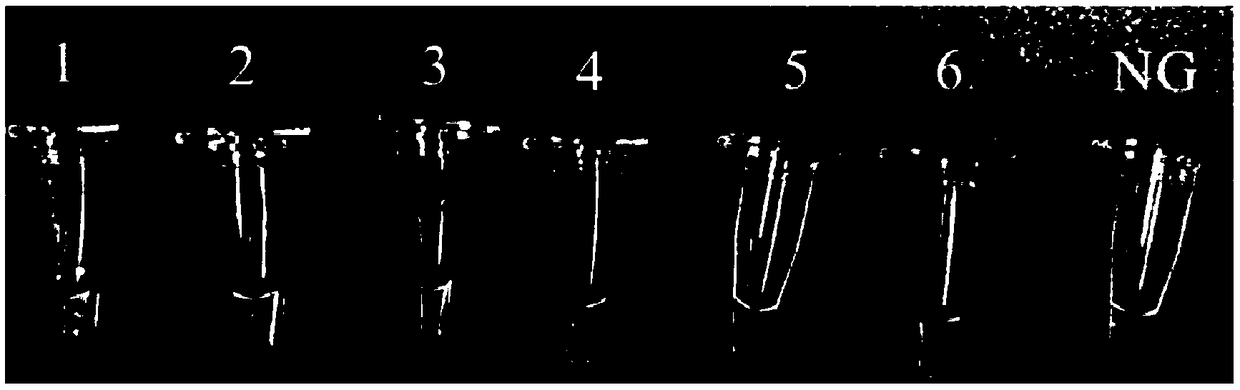
[0061] The genomic DNA of Escherichia coli O157:H7ATCC43895 and non-Escherichia coli was established according to the reaction system and conditions in Example 1. The polymerase helical reaction detection method was carried out, and the specificity test was carried out; wherein, the non-Escherichia coli was: Staphylococcus aureus ATCC23235, cheese Lactobacillus GBHM-21 (Bao Zhining. Research on identification, pilot scale preparation and fermentation technology of Lactobacillus casei GBHM-21 [D]. South China University of Technology, 2015.), Salmonella ATCC14028, Pseudomonas aeruginosa ATCC27853, Listeria monocytogenes ATCC19118. Escherichia coli O157:H7 genome is set as a positive control, and ultrapure water is a negative control (the results of the negative control are the same as figure 1 The results of the blank control in A), the results are as follows figure 2 shown. The reactio...
Embodiment 3
[0062] Embodiment 3PSR detects the sensitivity test of E.coli O157:H7
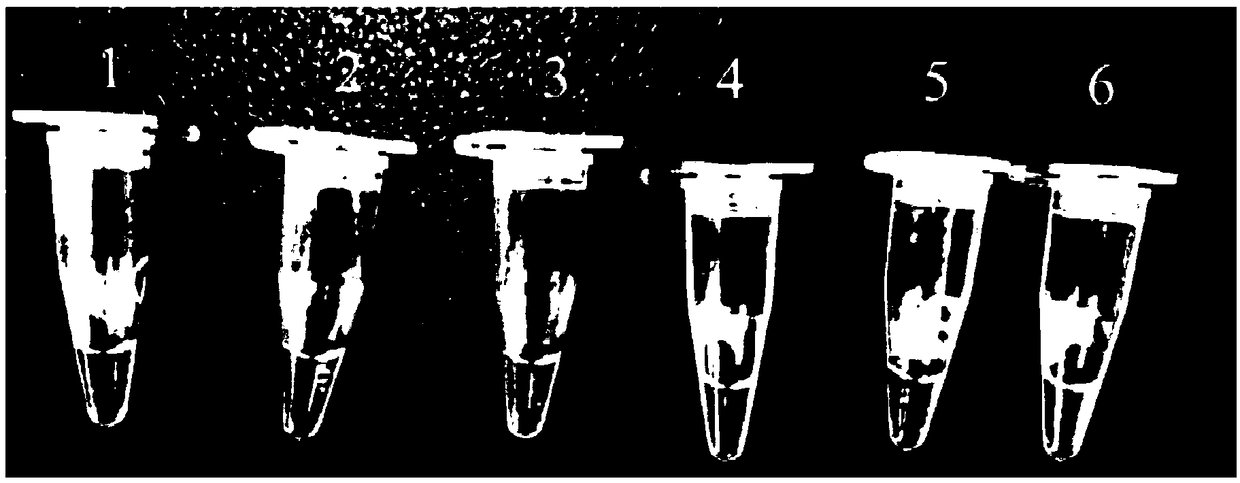
[0063] The genome of Escherichia coli O157:H7 was diluted 10-fold to 112ng / μL, 11.2ng / μL, 1.12ng / μL, 112pg / μL, 11.2pg / μL and 1.12pg / μL, and a negative control ( deionized water), construct the polymerase helical reaction amplification method according to the reaction system in Example 1, to determine the sensitivity of the detection method, the results are as follows image 3 shown. It can be seen from the figure that the reaction system turns green when the concentration of E. coli DNA in the sample is higher than 11.2pg / μL, showing a positive result. The results showed that the established E.coli O157 polymerase helical reaction method could detect 11.2pg / μL of Escherichia coli DNA in the sample.
[0064] Conclusion: From the above experimental results, it can be seen that the polymerase helical reaction amplification method has the following advantages over conventional PCR and fluorescent PCR:
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com