The Specific Sequence of Konjac Soft Rot and Its Detection Primers and Application
A soft rot fungus and konjac technology is applied in the field of konjac soft rot fungus specific sequences and detection primers, which can solve the problems of unsuitability for rapid application, non-specific amplification, and difficulty in distinguishing subspecies, etc., and achieve high-efficiency pathogen specific detection. The effect of target screening, good specificity, and avoiding false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Application of the specific sequence of Konjac soft rot pathogen (shown in SEQ ID NO.1) or primers designed for the sequence in the preparation of detection reagents for Konjac soft rot pathogen:
[0027] For the sequence shown in SEQ ID NO.1, LAMP primers were designed as follows:
[0028] F3: GCACAAGCTTGACTGCATAC
[0029] B3: TGGCGAGTTGTCCCCATAG
[0030] FIP: CGCCGTATCGGCACAGAAGAAAGCTTGGCGTTTCTCTCCA
[0031] BIP:GCCCCTCATCTCGCTGGCAATCATGTGCTTCCGGCAACAC
[0032] LF:AGAAACGGGATGGGGTGG
[0033] LB:GCCATCGAGCGTAGCGAA
[0034] The LAMP amplification system is as follows:
[0035]
[0036]
[0037] Bst 2.0 WarmStart DNA Polymerase was purchased from New England Biolabs.
[0038] The LAMP amplification procedure is as follows:
[0039] The reaction was carried out at 65°C for 40 minutes, and at 80°C for 5 minutes to stop the reaction.
[0040] Add 1000×SYBR Green I to the reaction product, the positive reaction shows green, indicating the existence of Konjac so...
Embodiment 2
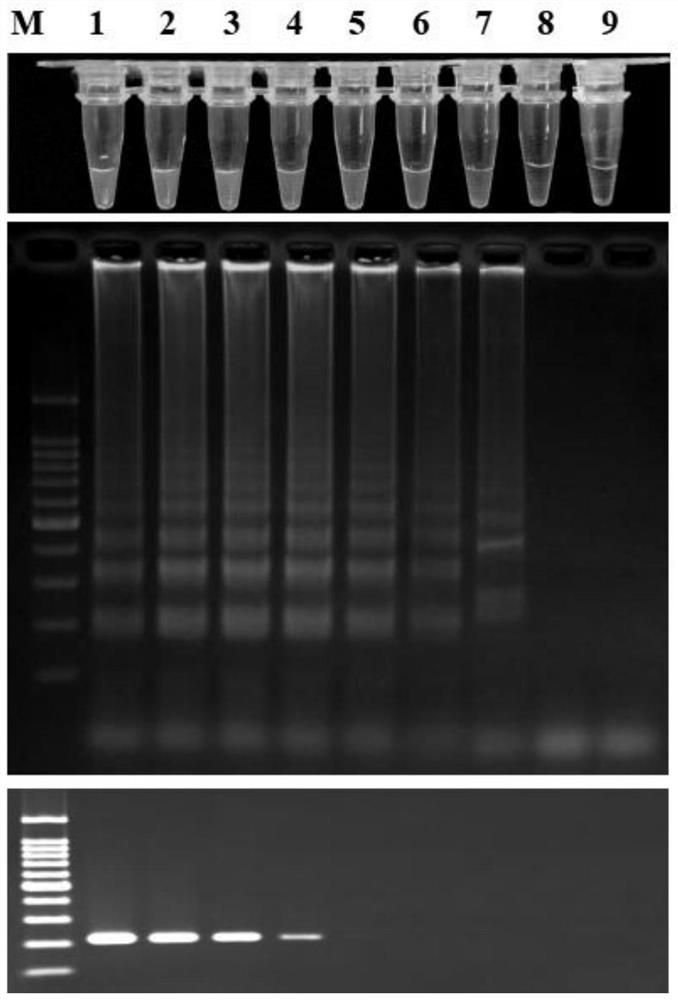
[0042] Konjac soft rot LAMP primer specificity detection and sensitivity detection:
[0043] Konjac soft rot fungus LAMP primer specificity detection:
[0044] Using the method described in Example 1, the applicant has carried out LAMP detection to 50 bacterial strains (Table 1), including the gene sequences of species or subspecies (such as: Pectobacterium carotovorum subsp. and Pectobacterium atrosepticum), Dickeya spp. (e.g. Dickeyadadantii and Dickeya zeae), Erwinia spp. (e.g. E. amylovora and E. tracheiphila), and other pathogenic bacteria (e.g. Ralstonia solanacearum and Pantoea ananatis), and Common bacteria in soil (such as: Agrobac terium tumefaciens and Bacillus subtilis), the results are shown in Table 1, the results show that only Pcc can be detected specifically, other bacterial strains can not be detected, and can accurately distinguish Pcc and its close relative subtilis Species were distinguished, indicating that this set of primers can specifically detect Kon...
Embodiment 3
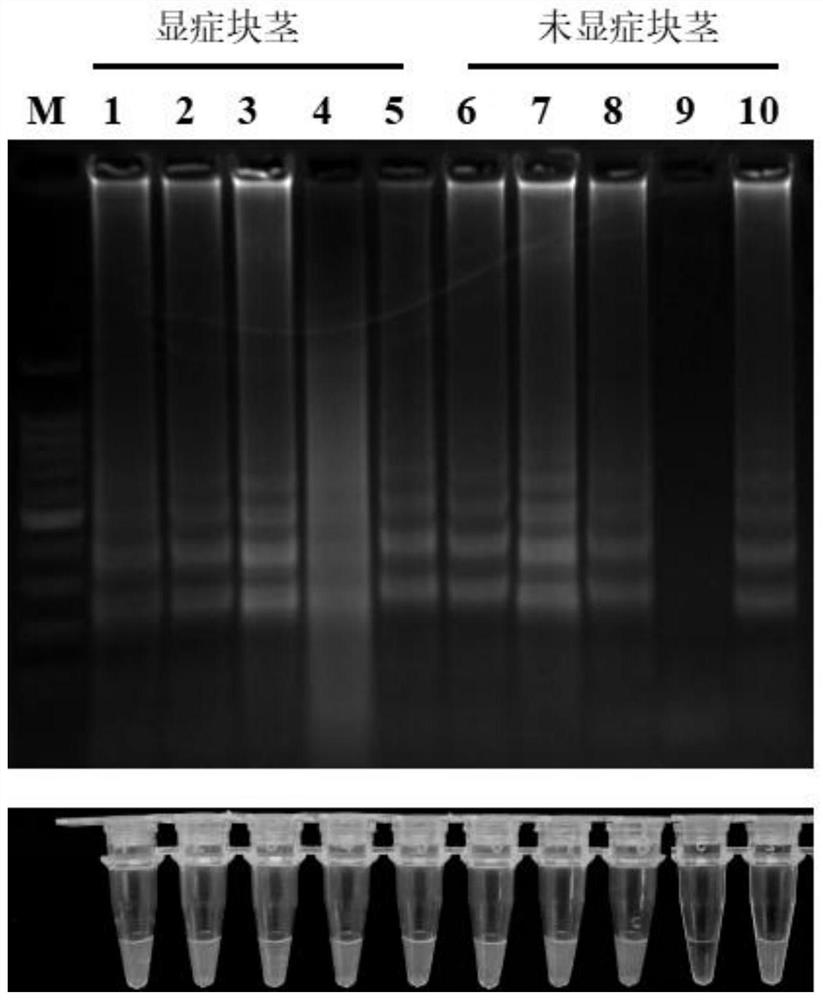
[0053] Field testing of Konjac soft rot fungus LAMP primers:
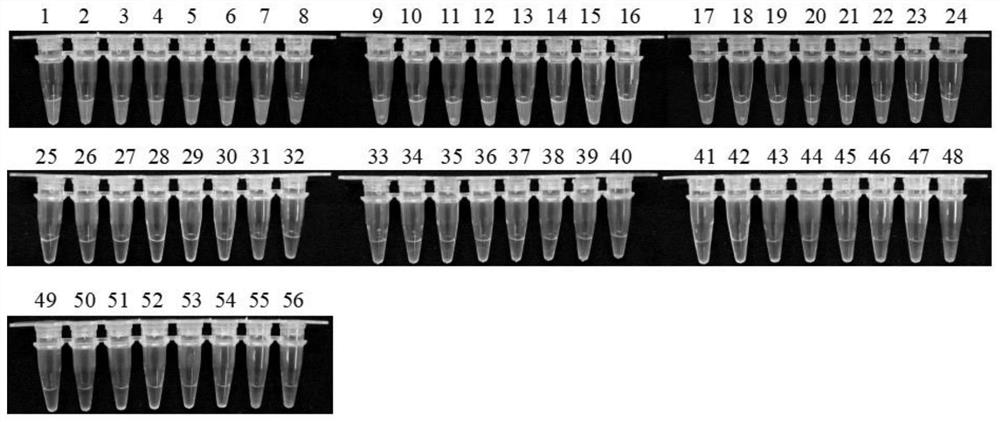
[0054] Using the method described in Example 1, 24 strains of Konjac soft rot fungi and other 31 negative control strains collected in the field were amplified using the LAMP primers of the present invention. The results showed that all 24 strains of Konjac soft rot fungi collected Detected, the LAMP amplification results of 31 negative control strains were all negative ( figure 1 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com