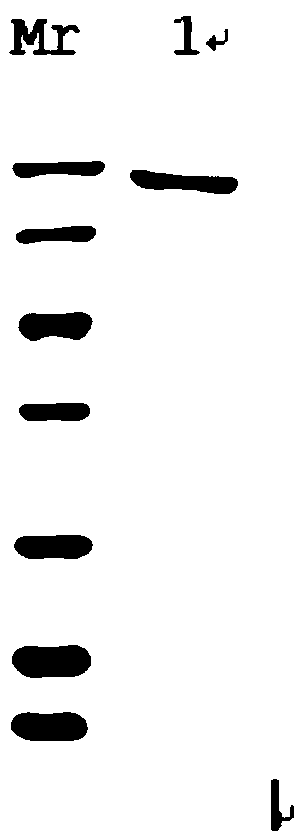Code error-prone DNA (Deoxyribonucleic Acid) polymerase and preparation method thereof
A technology of polymerase and coding, applied in the field of genetic engineering, can solve problems such as optimization and effort
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Sequencing and cloning of the full-length coding region of the gene:
[0025] The total DNA of Rhodococcus R04 was extracted using a commercially available bacterial genomic DNA extraction kit, sequenced by Huada Genomics, and the error-prone DNA polymerase sequence was obtained by blast comparison. As shown in NO: 2, it encodes 1123 nucleotides, with ATG as the start codon and TGA as the stop codon.
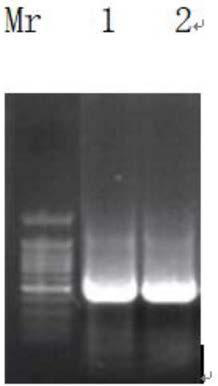
[0026] The error-prone DNA polymerase gene was amplified by PCR, and the amplification primers were SEQ ID NO: 3:GCGAATTCATGCAGTCGATCGTGCAGTT, SEQ ID NO: 4AGCGGCCGCGCGAAAGTCGCGTGAT. After the PCR product was detected by 1% agarose gel electrophoresis, it was cut under ultraviolet light containing For the gel block of the target band, use the agarose gel recovery kit to recover the target fragment, and the recovered product and the pET32a plasmid are subjected to EcoRI and NotI double digestion, and then electrophoresis to recover. The error-prone DNA polymeras...
Embodiment 2
[0027] Embodiment 2: Cultivation and induction of recombinant error-prone DNA polymerase strain
[0028] The expression strain was inoculated in 4 mL of LB liquid medium (Amp 100 g / mL) and cultured at 37°C for 16-20 h, then 1.25 mL of the culture solution was inoculated into a 250 mL shake flask, and cultured at 37°C for 4.5 h. Then add IPTG with a final concentration of 0.5mM for induction, and continue to cultivate for 16-20h.
Embodiment 3
[0029] Example 3: Extraction of error-prone DNA polymerase
[0030] Centrifuge at 12000rpm for 10min at 4°C to collect the bacteria, resuspend in 50mL 20mM pH 8.0 phosphate buffer, centrifuge, collect the precipitate, resuspend in 20mL 20mM pH 8.0 phosphate buffer, add 8mg of lysozyme to it to make the final concentration reach 0.4mg / mL. After placing at 37°C for 30 minutes, centrifuge at 10,000 rpm for 20 minutes, and collect the supernatant, which is the error-prone DNA polymerase extract.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com