Primers and probe for detecting hog cholera virus based on a digital PCR technology, a kit and a method thereof
A swine fever virus and technical detection technology, applied in the field of molecular biology, can solve the problems of absolute quantitative detection of swine fever virus, achieve broad application prospects and industrialization prospects, not easily affected, and accurately detect the effect of virus content
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044]Embodiment 1 is based on the establishment of the kit for the absolute quantitative detection of swine fever virus by digital PCR technology
[0045] A kit for the absolute quantitative detection of classical swine fever virus based on digital PCR technology, including a primer set, 2×RT-ddPCR Supermix, RNase-free distilled water, virus total RNA extraction reagent, positive control and negative control.
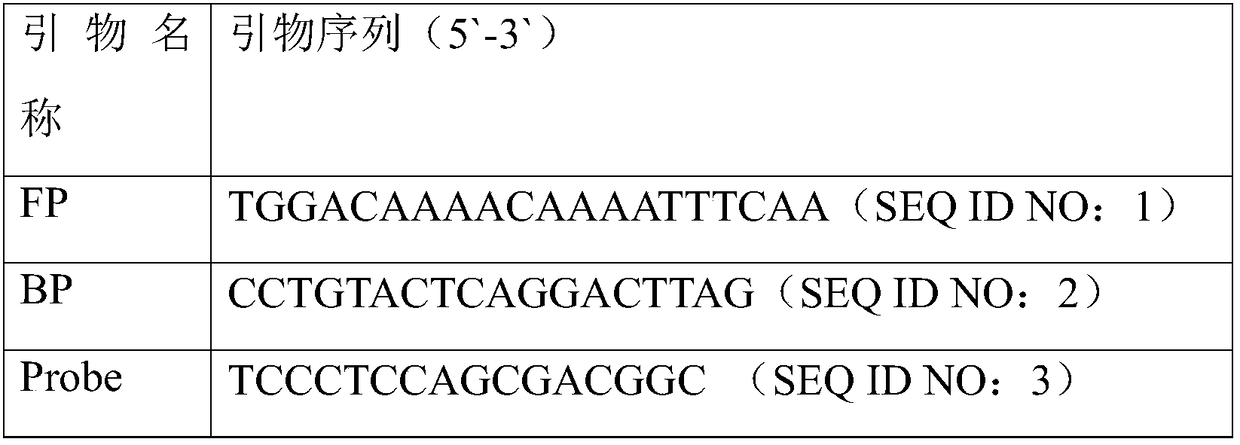
[0046] (1) Design of primers for digital PCR amplification: primers were designed with the specific conserved sequence of classical swine fever virus as the target gene. The primer sequences are listed in Table 1.
[0047] Table 1 Primer sequence list
[0048]
[0049] (2) The molar ratio of FP primer, BP primer and probe in the primer set is 3:3:2.
[0050] (3) 2×RT-ddPCR Supermix contains: 2×One-step RT-ddPCR Supermix and 25mMManganous acetate.
[0051] (4) The positive control is the cell culture of classical swine fever virus, and the negative control is deio...
Embodiment 2
[0053] Embodiment 2 Absolute Quantitative Detection Method Based on Digital PCR Technology to Detect Classical Swine Fever Virus
[0054] The method utilizing the test kit of embodiment 1 to detect classical swine fever virus may further comprise the steps:
[0055] (1) Extraction of viral RNA:
[0056] 1) Take the sample to be tested and the positive control, add 600ul lysate A respectively, vortex and mix for 20 seconds, and let stand at room temperature for 10 minutes;
[0057] 2) Transfer the mixture to an adsorption column and centrifuge at 12,000×g for 30-60 seconds;
[0058] 3) Discard the liquid in the collection tube, add 500ul washing solution B to the adsorption column, and centrifuge at 12,000×g for 30-60 seconds;
[0059] 4) Discard the liquid in the collection tube, add 500ul washing solution C to the adsorption column, and centrifuge at 12,000×g for 30-60 seconds;
[0060] 5) Discard the liquid in the collection tube, and centrifuge at 12,000×g for 2 minutes ...
Embodiment 3
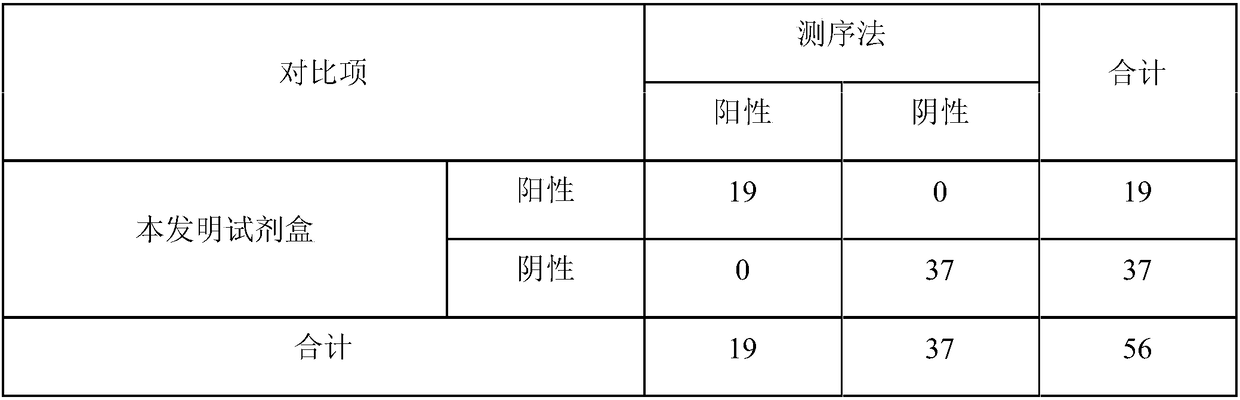
[0079] Example 3 specificity verification
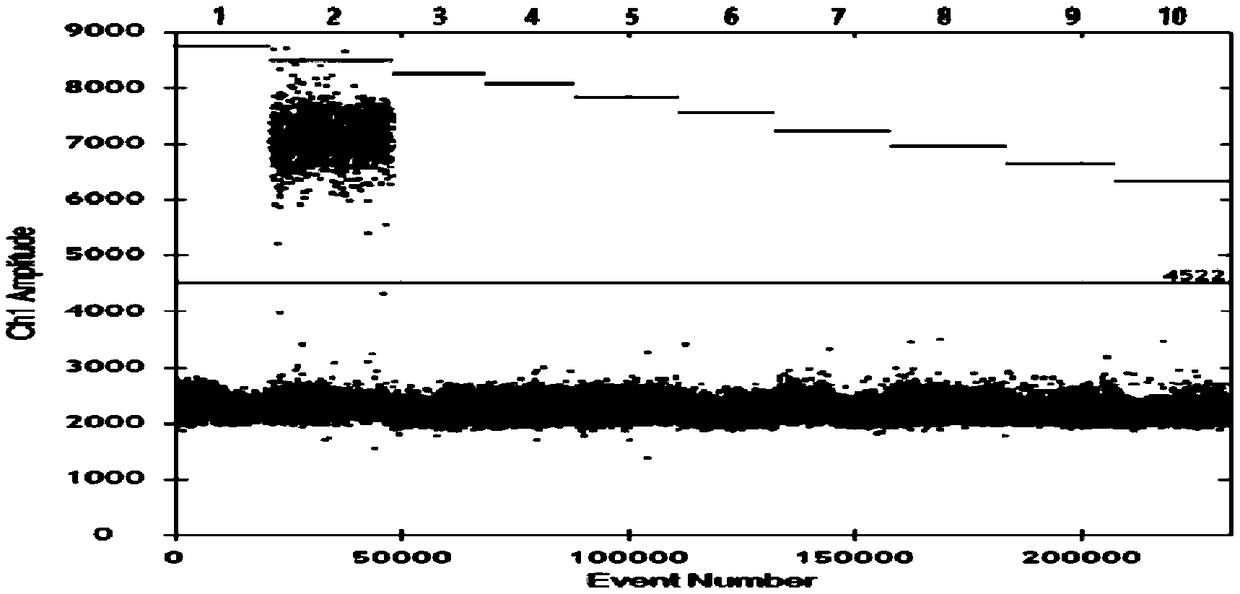
[0080] The serum samples of healthy pigs are detected respectively with the kit of the present invention, and clinically obtained pseudorabies virus, porcine respiratory and reproductive syndrome American type virus, porcine epidemic diarrhea virus, porcine circovirus, Haemophilus parasuis, Streptococcus suis, There are a total of 8 samples of Actinobacillus pleuropneumoniae samples, all samples have been verified by sequencing method, and the test results can be found in figure 1 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com