Kit for identifying lung nodules and/or lung cancer status and application thereof
A kit and technology for pulmonary nodules, applied in the field of kits for identifying pulmonary nodules and/or lung cancer status, to achieve the effects of improving sensitivity and specificity, rapid diagnosis and evaluation, and ensuring correctness and reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1: DNA Extraction
[0062] DNA Extraction Reagent consists of Lysis Buffer, Binding Buffer, Washing Buffer and Elution Buffer. Lysis buffer consists of protein denaturants, detergents, pH buffers and nuclease inhibitors. Binding buffers consist of protein denaturants and pH buffers. The cleaning buffer is divided into cleaning buffer A and cleaning buffer B, and cleaning buffer A is composed of protein denaturant, nuclease inhibitor, detergent, pH buffer and ethanol; cleaning buffer B is composed of nuclease inhibitor, pH buffer and ethanol composition. The elution buffer consists of nuclease inhibitors and pH buffers. The protein denaturant is: guanidine hydrochloride; the detergent is: Tween20; the pH buffer is: Tris-HCl; the nuclease inhibitor is: EDTA.
[0063] In this embodiment, the plasma samples of lung cancer patients are taken as an example to extract plasma DNA. The extraction method comprises the following steps:
[0064] (1) Take 1ml of plasma...
Embodiment 2
[0076] Example 2: Bisulfite Treatment of DNA
[0077] Bisulfite treatment of DNA is to use bisulfite reagent to treat the extracted DNA sample. The bisulfite reagent is composed of bisulfite buffer and protection buffer; the bisulfite buffer is bisulfite A mixture of sodium and water; the protection buffer is a mixture of hydroquinone, an oxygen free radical scavenger, and water.
[0078] In this embodiment, the DNA extracted in Example 1 is used as the processing object, and the DNA is treated with bisulfite, and the specific steps include:
[0079] (1) Preparation of bisulfite buffer: weigh 1g of sodium bisulfite powder, add water to prepare 3M buffer;
[0080] (2) Preparation of protection buffer: weigh 1g of hydroquinone reagent, add water to prepare 0.5M protection buffer;
[0081] (3) Mix 100 μl DNA solution, 200 μl bisulfite buffer solution and 50 μl protection solution, shake and mix evenly;
[0082] (4) Thermal cycle: 95°C for 5 minutes, 80°C for 60 minutes, 4°C fo...
Embodiment 3
[0093] Example 3: Detection of DNA methylation by real-time fluorescent PCR and verification of primer sets
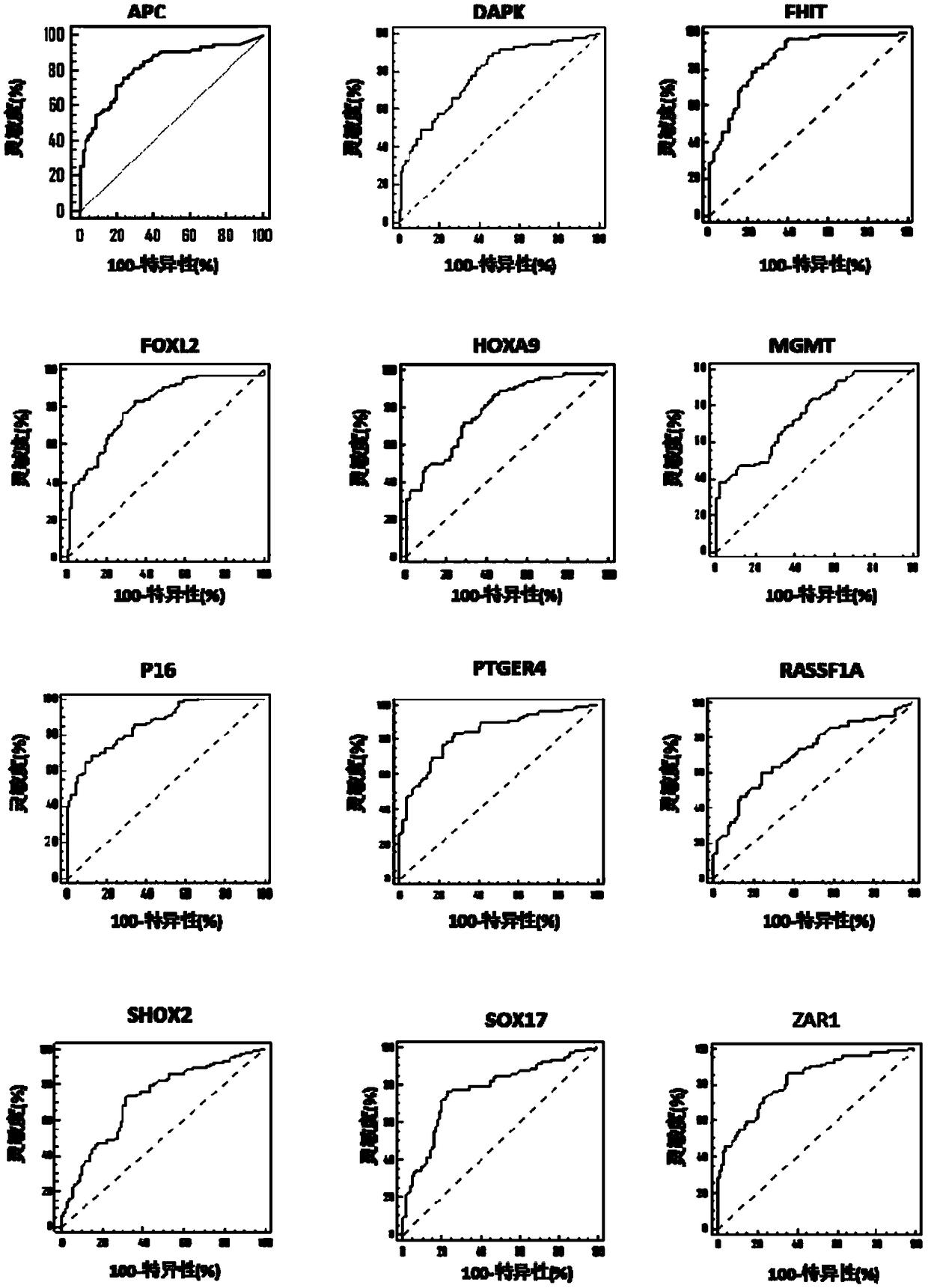
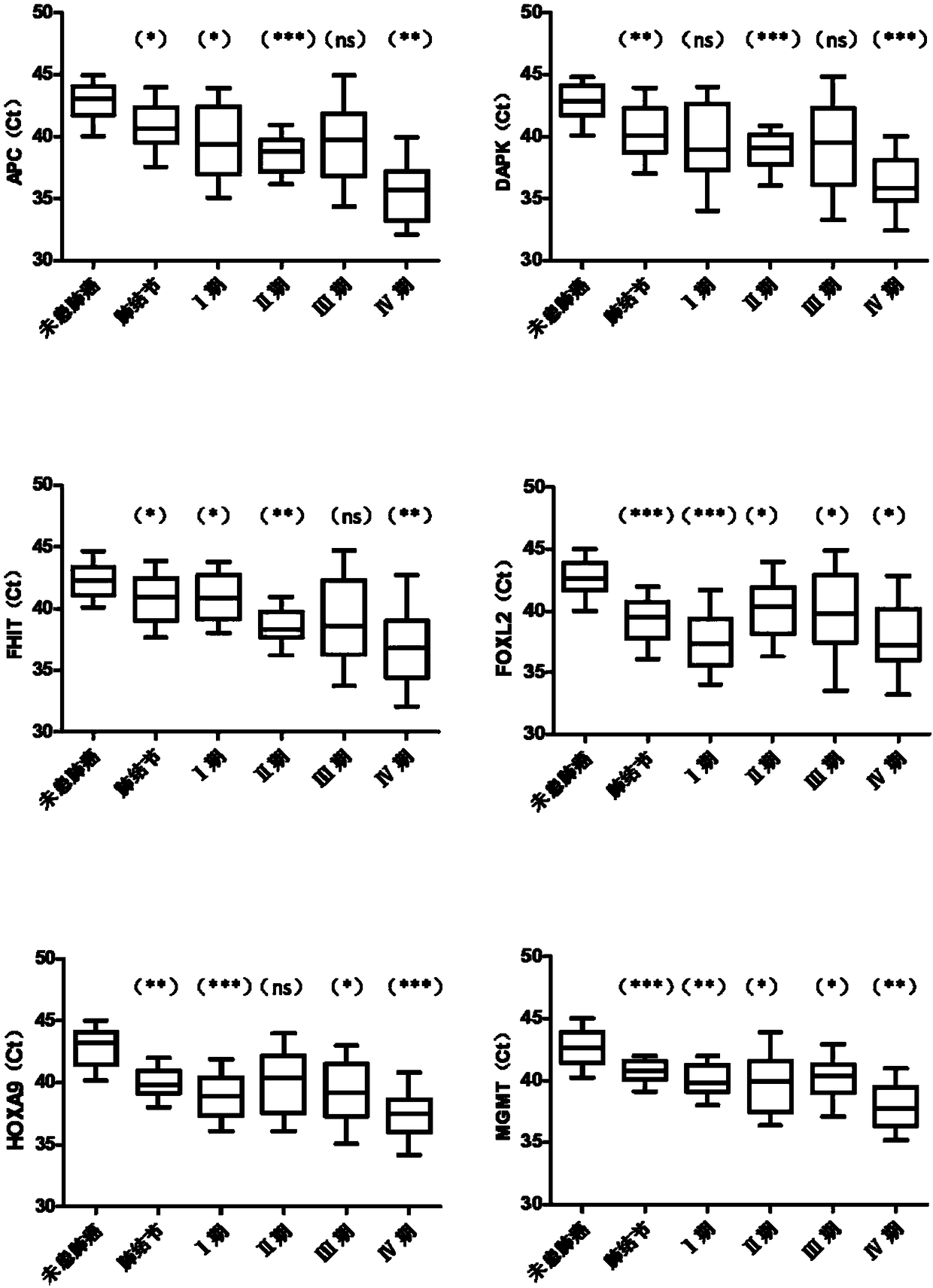
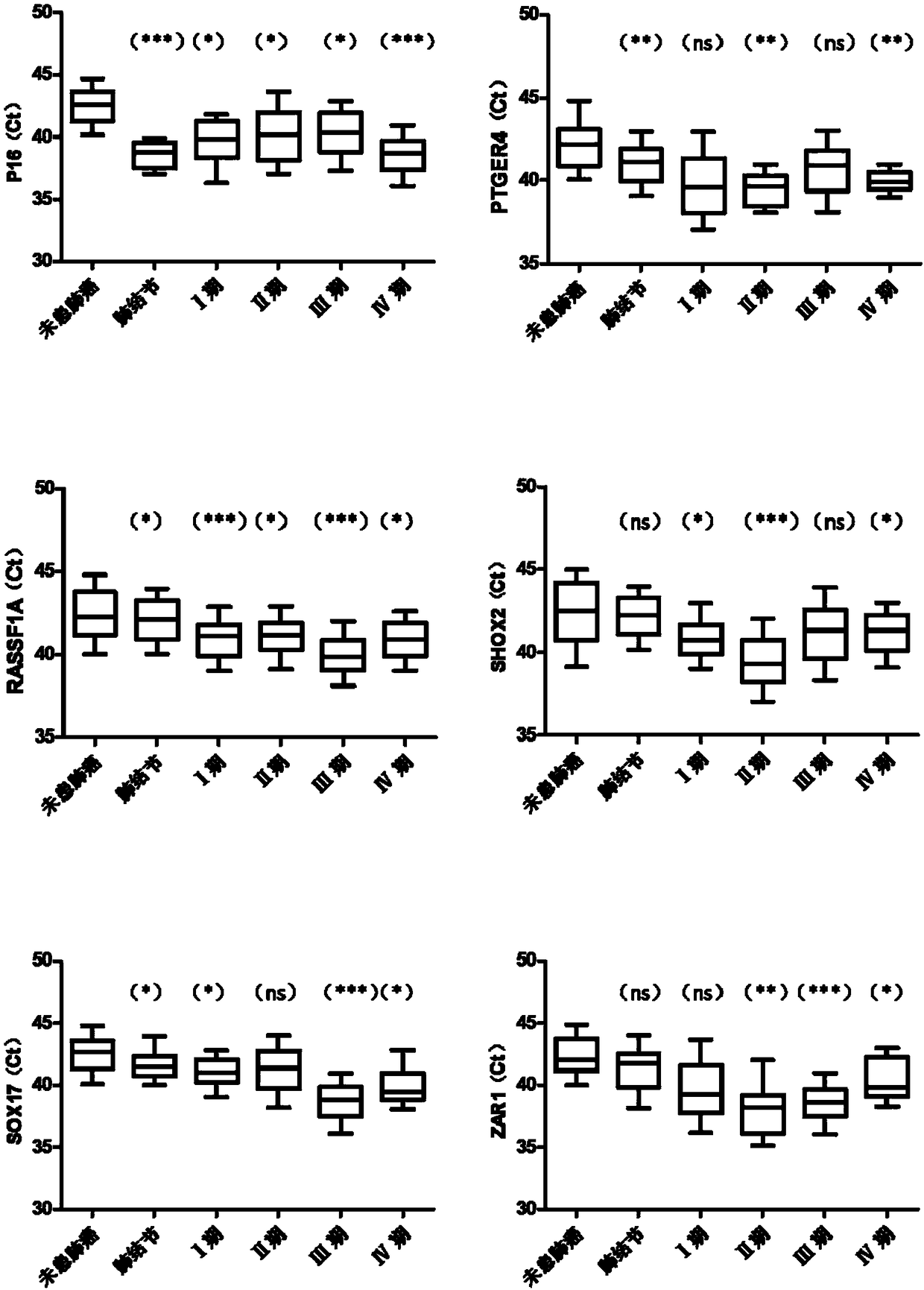
[0094] In this embodiment, real-time fluorescent PCR is used as an example to detect the methylation level of biomarker genes. The detected genes were APC, DAPK, FHIT, FOXL2, HOXA9, MGMT, P16, PTGER4, RASSF1A, SHOX2, SOX17 and ZAR1 genes, and the internal reference gene was ACTB. In this example, the DNA treated with bisulfite in Example 2 was used as a template for real-time fluorescent PCR amplification. The DNA samples to be tested, the negative quality control, the positive quality control, and the no-template control were all tested in triplicate holes.
[0095] For APC, DAPK, FHIT, FOXL2, HOXA9, MGMT, P16, PTGER4, RASSF1A, SHOX2, SOX17 and ZAR1 genes, many sets of primer and probe combinations can be designed, and the performance of each set of probe-primer combinations may vary. difference, so it needs to be verified by experiments. In this example, the prime...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com