A method for increasing the expression of chitinase
A chitin enzyme and expression technology, which is applied in the fields of enzyme engineering and microbial engineering, can solve problems such as complex operation, loss of enzyme activity, and difficulty in screening, and achieve great application prospects and high-efficiency secretion and expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1: Construction of recombinant bacteria
[0048] Specific steps are as follows:
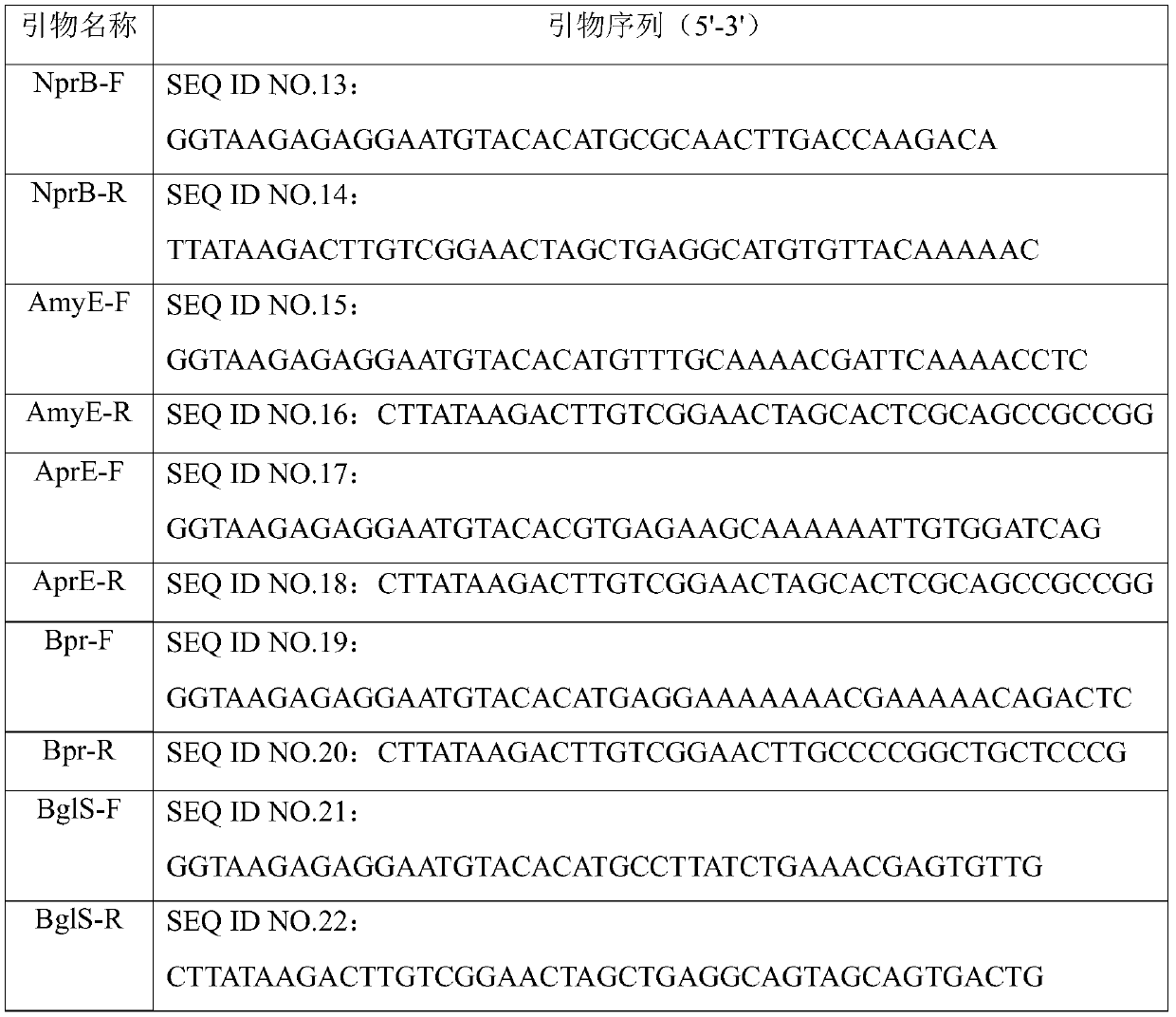
[0049] (1) Using Bacillus subtilis 168 as a template, respectively using NprB-F, NprB-R, AmyE-F, AmyE-R, AprE-F, AprE-R, Bpr-F, Bpr-R, BglS-F, BglS -R, Epr-F, Epr-R, LipA-F, LipA-R, Vpr-F, Vpr-R, YclQ-F, YclQ-R, YweA-F, YweA-R as forward and reverse primers (see table 1-2), 10 signal peptide fragments were amplified by PCR: NprB, AmyE, AprE, BglS, Bpr, Epr, LipA, Vpr, YweA, YclQ; the PCR reaction conditions were: 98°C for 3min, 30 cycles (98 ℃30s, 55℃30s, 72℃30s), 72℃5min;
[0050] (2) Use pP43NMK-chisb (the chisb gene was synthesized by Wuxi Tianlin Biological Co., Ltd. and provided the constructed plasmid) as a template, and p43-F and p43-R as forward and reverse primers (see Table 1-2 ), the linearized carrier fragment containing the chitinase gene whose N-terminus is deleted from its own signal peptide gene was amplified by PCR of the whole plasmid; 72℃4min30s), 72℃5min;...
Embodiment 2
[0065] Embodiment 2: the verification of high-yielding chitinase recombinant bacteria
[0066] Specific steps are as follows:
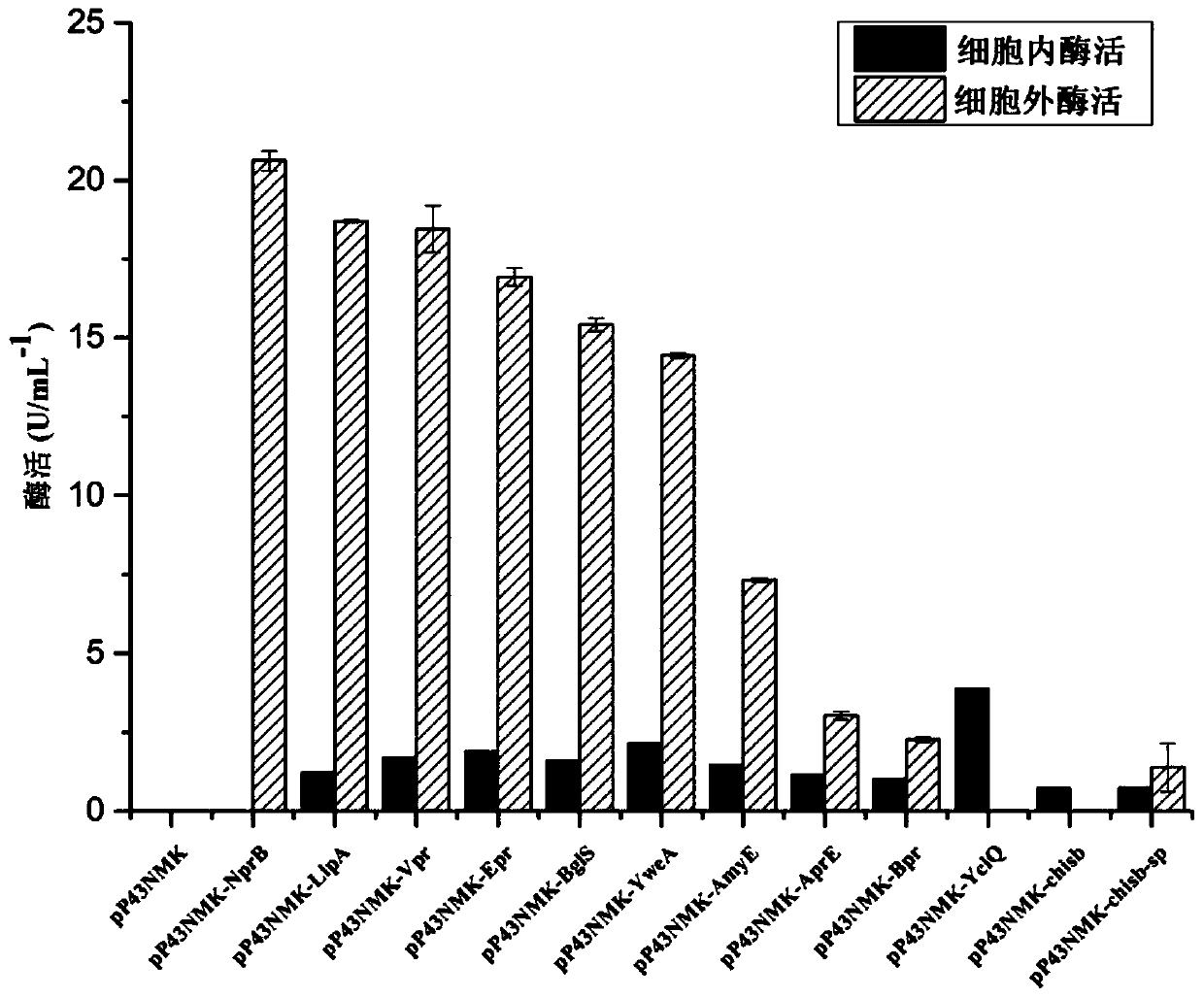
[0067] The recombinant plasmids pP43NMK-NprB, pP43NMK-AmyE, pP43NMK-AprE, pP43NMK-BglS, pP43NMK-Bpr, pP43NMK-Epr, pP43NMK-LipA, pP43NMK-Vpr, pP43NMK-LipA, pP43NMK-Vpr, pP43NMK-YweA and pP43NMK-YclQ were respectively transformed into Bacillus subtilis WB600, and the selected transformants were inoculated into LB liquid medium, cultured at 37°C for 8 hours, then transferred to TB medium with an inoculum size of 2%, cultured for 12 hours, collected and fermented The supernatant of the fermentation liquid was used to detect the enzyme activity of the fermentation supernatant; at the same time, the cell wall was broken by ultrasonic disruption, and the enzyme activity in the cell was detected.
[0068] The result is as figure 1 Shown (with empty plasmid pP43NMK, recombinant plasmid pP43NMK-chisb, recombinant plasmid pP43NMK-chisb-sp as control).
[0069...
Embodiment 3
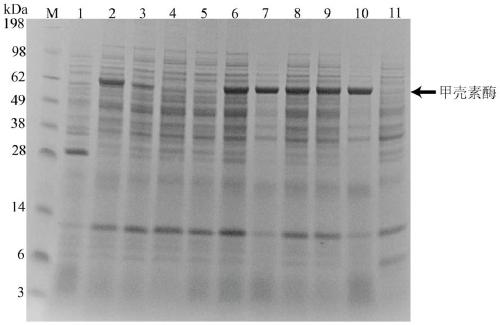
[0070] Example 3: Protein electrophoresis verification of chitinase production strains expressing different signal peptides
[0071] The fermentation supernatant in Example 2 was taken, and the recombinant strains containing different signal peptides were subjected to protein sample treatment. System: 30 μL fermentation supernatant, 10 μL 4× protein loading buffer, 99 ° C, 10 min, and then protein electrophoresis. Through dyeing, decolorization and other processes, the results are as follows figure 2 shown.
[0072] The results showed that the bands of the recombinant strains fused with different signal peptides were all obviously thicker, indicating that the yield of chitinase was significantly increased. At the same time, it was found that the recombinant strain pP43NMK-YclQ fused with the YclQ signal peptide had no protein band display, indicating that it had no protein bands. The ability to secrete chitinase extracellularly. The recombinant strain pP43NMK-NprB protein b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com