A detection method for single molecular protein
A detection method and single-molecule technology, applied in the field of protein detection, can solve the problems of inability to meet the requirements for detection of proteins with low expression abundance and low detection sensitivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] 1. Laser confocal detection system ( figure 1 with figure 2 )
[0081] The laser confocal detection system includes a microfluidic chip A, a point light source B, an imaging unit C, a scanning lens D and a three-dimensional droplet imaging unit.
[0082] The point light source B adopts a laser or an LED lamp to form a point light source and project it onto the droplet.
[0083] The imaging unit C is an EMCCD photon detector, and the imaging unit C collects and detects the fluorescent signals processed by the four-channel fluorescent filter F and the lens G, and can use different fluorescent channels to independently synthesize the characteristics of the droplet containing the fluorescent signal Numerical imaging of droplet shape and fluorescence intensity °C.
[0084] 2. Microfluidic chip
[0085] The preparation of the hot-pressing mold, using the nickel mold to prepare the upper structure, see the detailed structure figure 1, wherein the width of the droplet for...
Embodiment 2
[0094] Example 2: Detection of GPC1 by direct fluorescence method
[0095] Preparation of multilayer droplets:
[0096] (1) Prepare the oil phase. Prepare the oil phase according to the following proportions (both by mass): 50g mineral oil + 50g n-tetradecane + 1.5g emulsifier EM90 + 1.5g TritonX-100, and use it for later use after ultrasonic degassing.
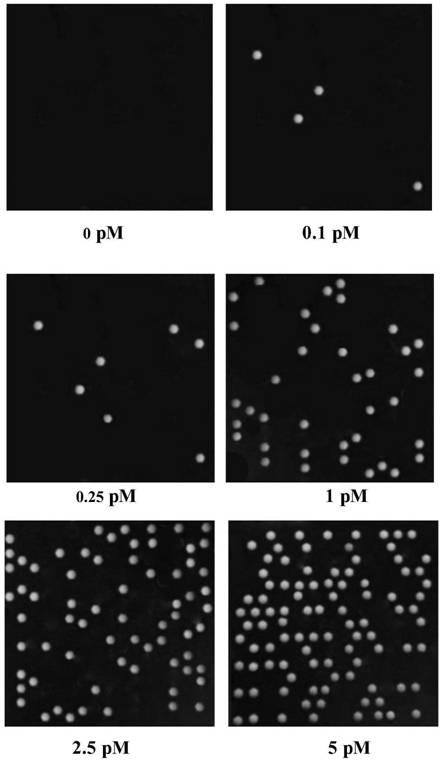
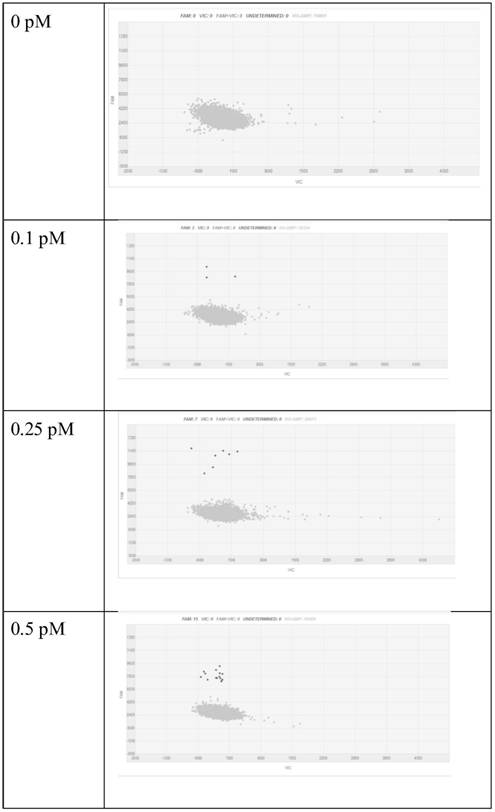
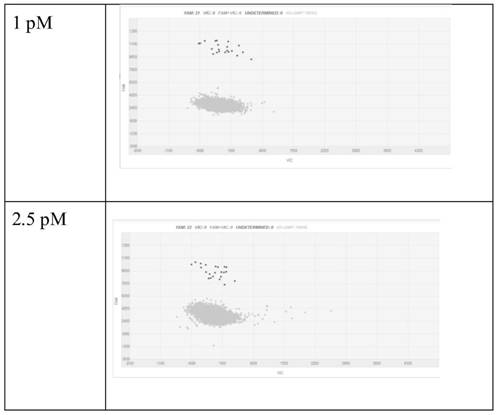
[0097] (2) Prepare the water phase. Prepare different concentrations of GPC1 protein standards (abcam, ab114484), that is, use the standard diluent to dilute the original GPC1 protein standard, and the final GPC1 protein concentrations are 0, 0.1, 0.25, 0.5, 1.0, 2.5, 5 and 10PM, respectively. , the final volume is not less than 200 μl. Use Merck’s magnetic beads to couple GPC1 capture antibody (abcam, ab55971) 6 μL GPC1 protein 200 μl in the capture solution, incubate at 37 ° C for 1 h, wash the supernatant, and then add FITC-labeled detection antibody (Invitrogen, Cat#62-8411) 200 μl , the detection antibody was diluted...
Embodiment 3
[0101] Example 3: Detection of GPC1 by HRP enzyme labeling method
[0102] Preparation of multilayer droplets:
[0103] (1) Prepare the oil phase. Prepare the oil phase according to the following proportions (both by mass): 50g mineral oil + 50g n-tetradecane + 1.5g emulsifier EM90 + 1.5g TritonX-100, and use it for later use after ultrasonic degassing.
[0104] (2) Prepare the water phase. Prepare different concentrations of GPC1 protein standards (abcam, ab114484), that is, use the standard diluent to serially dilute the original GPC1 protein standard, to the final GPC1 protein concentration of 0, 0.1, 0.25, 0.5, 1.0, 2.5, 5 and 10pM The final volume is not less than 200 μl. 200 μl of GPC1 protein in 6 μl capture solution of Merck’s magnetic bead-coupled GPC1 capture antibody (abcam, ab55971), incubated at 37°C for 1 h, washed the supernatant, and then added 200 μl of HRP-labeled detection antibody (Invitrogen, Cat#31430). The detection antibody was diluted with PBS at a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com