A kind of rhizobia famb126 and its application
A FAMB126, rhizobia technology, applied in the field of microorganisms, to achieve the effect of improving total nitrogen content, high nodulation rate and strong nitrogen fixation ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1 Isolation and purification of Rhizobium FAMB126
[0023] (1) During the flowering period of normal growth of peanuts, take fresh whole peanut roots, take fresh, mature and large plump nodules of peanuts from the roots, rinse with water, and absorb surface water with filter paper.
[0024] (2) Put it in 95% ethanol with volume fraction for 3~5 minutes, take it out and rinse with sterile water for 5~6 times, then put in 1g / L HgCl 2 Sterilize for 3 to 5 minutes, take out and rinse with sterile water 5 to 6 times.
[0025] (3) Cut into two halves on a flame-sterilized glass slide, clamp half of the tumor with sterile tweezers, scribe the incision facing the surface of the YMA (Table 1) medium, invert it and incubate at 28°C.
[0026] (4) After the bacteria grow, use a sterile toothpick to scrape a small amount of rhizobia colonies from the plate, dilute with sterile water, and streak the culture on the plate again. After 3 days, observe the colony situation, until 15 Day...
Embodiment 2
[0033] Example 2 Rhizobium 16s rDNA sequence sequencing
[0034] PCR specific amplification of Rhizobium monoclonal bacteria liquid, the primers are V3-V4-F and V4-V5-R, the forward primer V3-V4-F is 5′-GWATTACCGCGGCKGCTG-3′; the reverse primer V4 -V5-R is 5'-CCGTCAATTCMTTTRAGTTT-3', and the enzyme is 2×star Mix. The PCR amplification products are detected by electrophoresis imaging technology to observe whether they have bands, and the remaining PCR amplification products are used for sequence determination. The sequencing result is shown in SEQ ID NO:1. The PCR reaction system is as follows:
[0035] Table 2 16s rDNA 2×starMix enzyme reaction system
[0036]
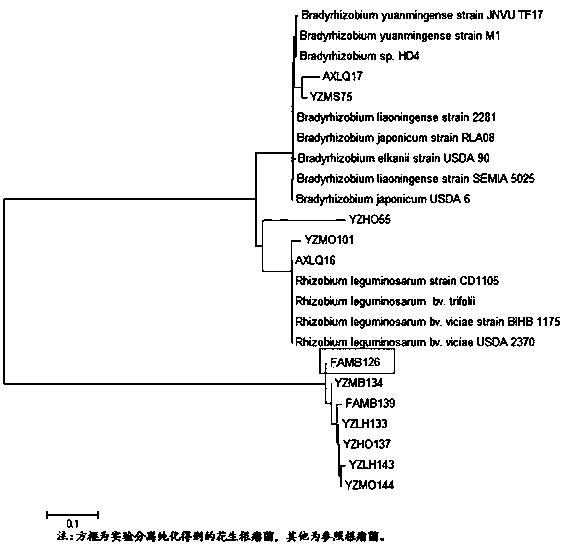
[0037] Obtain 12 reference strain sequences from the NCBI (GenBank) database, and use the software BioEdit and MEGA6 to analyze the isolated strains and reference strains. 16s rDNA partial sequence analysis to construct a phylogenetic tree of the isolated strain and the reference strain (attached figure 2 ). Therefore, ...
Embodiment 3
[0038] Example 3 Tie back test
[0039] 1. Strain culture:
[0040] The test strains stored at -80°C were transferred to YMA liquid culture and cultivated to the logarithmic phase. The growth of the strains was detected with a nucleic acid protein analyzer, and the bacterial content was calculated using the OD value. The formula of YMA (Yeast Mannnitol Agar) liquid medium is: Weigh 10 g of mannitol, MgSO 4 ∙7H 2 O 0.2g, NaCl 0.1 g, yeast powder 3 g, K 2 HPO 4 0.25g, KH 2 PO 4 0.25 g, CaCO 3 3 g (added when storing), 15 g of agar dissolved in 1 L of pure water.
[0041] 2. Sterilization treatment:
[0042] Wrap the 2.5L plastic buckets, beakers, tweezers, petri dishes, filter paper, and glass rods required for the test, and put the vermiculite in a fresh-keeping bag, sterilize at 121°C for 30 min. YMA liquid culture medium and distilled water are bottled and sterilized at 121°C for 20 min.
[0043] Put the seeds of peanuts into 1g / L HgCl 2 Sterilize the epidermis in the middle for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com