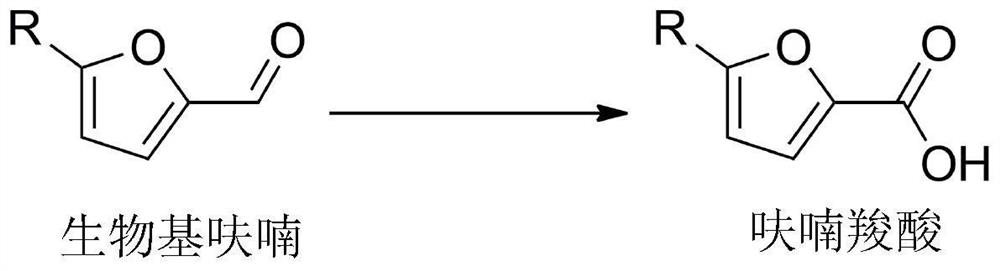
Aldehyde dehydrogenase and its gene, construction of recombinant bacteria and its application in the synthesis of furan carboxylic acid
An aldehyde dehydrogenase and furan carboxylic acid technology, which is applied in the fields of genetic engineering technology and biocatalysis, can solve the problems of reduced conversion efficiency, prolonged reaction time and the like, and achieves the effects of simple production process, high production efficiency and mild production conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Mining of Aldehyde Dehydrogenase Encoding Genes
[0044] In order to excavate the key enzyme in the genome of Comamonas testosteroni SC1588 that can catalyze the oxidation of bio-based furan to furan carboxylic acid, Shenzhen Huada Gene Technology Service Co., Ltd. was entrusted to sequence the draft genome of Comamonas testosteroni SC1588, according to According to the functional annotation of the predicted coding sequence, five aldehyde dehydrogenase genes were mined from the genome, namely, CtCALDH1 gene, CtCALDH2 gene, CtVDH1 gene, CtVDH2 gene and CtSAPDH gene.
Embodiment 2
[0046] Construction of recombinant bacteria
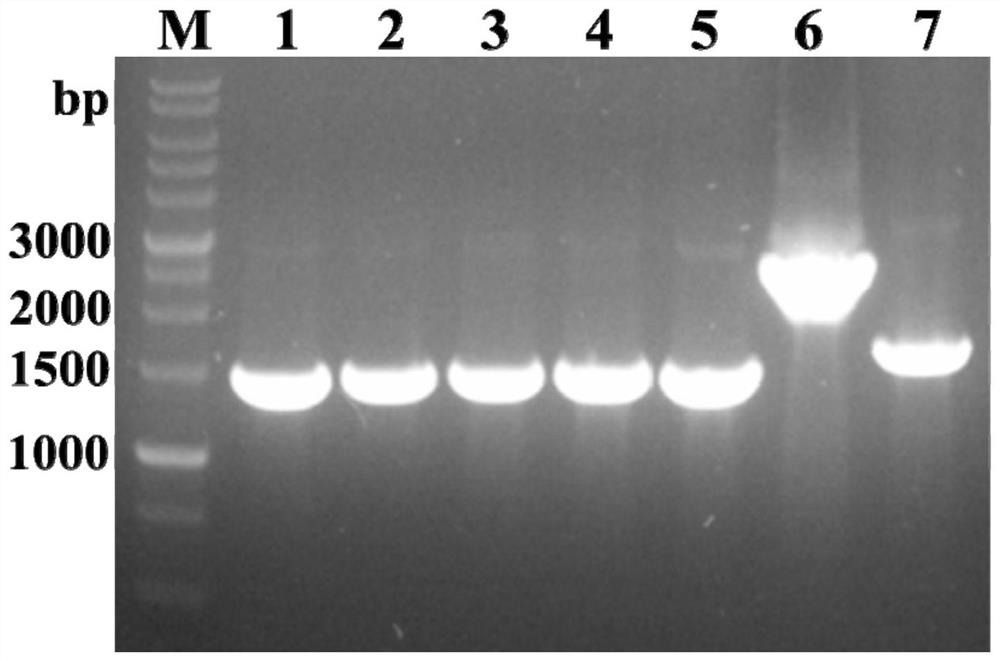
[0047] (1) Using Comamonas testosteroni SC1588 genomic DNA as a template, the full-length sequences of CtCALDH1, CtCALDH2, CtVDH1, CtVDH2 and CtSAPDH genes were amplified by designing specific primers;
[0048] (2) connecting the aldehyde dehydrogenase gene to the expression vector pET-28a to obtain a recombinant plasmid;
[0049] (3) Transform the recombinant plasmid verified by sequencing into the expression host E.coli BL21(DE3), and obtain recombinant bacteria E.coli BL21(DE3)_CtCALDH1, E.coli BL21(DE3)_CtCALDH2, E.coli BL21(DE3)_CtCALDH2, E. .coli BL21(DE3)_CtVDH1, E.coli BL21(DE3)_CtVDH2 and E.coli BL21(DE3)_CtSAPDH.
[0050] Table 1 Primer Information
[0051] Primer name Primer sequence (5'-3') CtCALDH1 upstream primer CAGGATCCATGACCCAGACGAATTCGTCCATC CtCALDH1 downstream primer ATGAAGCTTGGGAGTGACTGCGGCCTGCA CtCALDH2 upstream primer CGGGATCCATGAACTACATGGACTTGCACCGCA CtCALDH2 downstream...
Embodiment 3
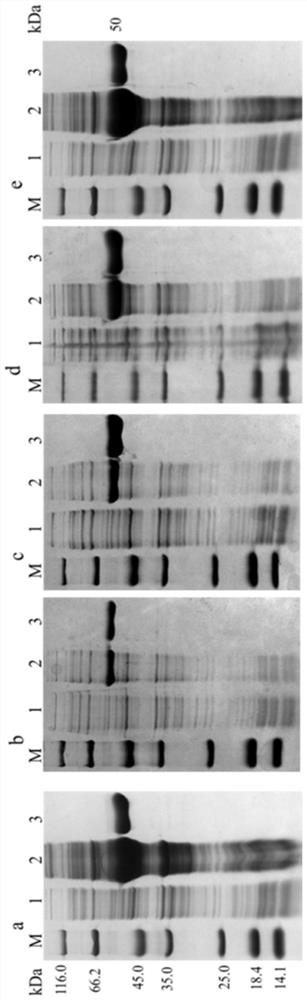
[0053] Inducible expression of aldehyde dehydrogenase
[0054] Inoculate the recombinant bacteria obtained in Example 2 into 30 mL of LB liquid medium (tryptone 10 g / L, yeast extract 5 g / L, sodium chloride 10 g / L, pH 7.2) containing 50 μg / mL kanamycin , cultivated at 37°C and 180rpm for 12h. Then, transfer 1% of the inoculum to 100 mL LB medium containing 50 μg / mL kanamycin, and culture at 37° C. and 180 rpm. OD of bacterial solution 600 When it reached 0.6-0.8, IPTG was added to make the final concentration 0.1 mM, and placed at 18° C. and 160 rpm for 20 h of induction culture. After the cultivation, the bacterial cells were collected by centrifugation at 8000 rpm and 4° C. for 5 min, and the cells were washed twice with 0.85% physiological saline.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com