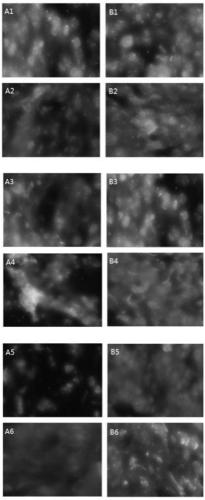
Preparation method of FISH probe of TOP2A gene
A probe and gene technology, applied in the field of FISH probe preparation, can solve problems such as affecting the accuracy of detection, increasing false positive and false negative signals, and reducing detection sensitivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0115] A method for preparing a FISH probe library of TOP2A gene includes the following steps:
[0116] (1) Prepare probe 1A and probe 1B;
[0117] (2) After cutting and interrupting probe 1A and probe 1B obtained in step (1) respectively, using Taq DNA polymerase for end-filling and 3'-end dA tailing to obtain probes 2A and 2B, respectively;
[0118] The interrupted system of enzyme digestion is:
[0119] Reagent
Add amount
10×CutSmart Buffer
5μL
2μL
Probe 1A or 1B (1ng / μL)
40μL
Taq DNA polymerase (5U / μL)
0.5μL
10mM dNTP
1μL
ddH 2 O
1.5μL
[0120] The reaction conditions for digestion interruption are: digestion at 37°C for 30 minutes, reaction at 72°C for 20 minutes, and reaction at 80°C for 20 minutes;
[0121] (3) Add linkers to probes 2A and 2B obtained in step (2) to obtain probes 3A and 3B, respectively;
[0122] The ligation reaction system is:
[0123] Reagent
Add amount
10×T4 DNA ligase buffer
10μL
T4DNA Ligase (400U / μL)
4μ...
Embodiment 7
[0128] The conditions of the ligation reaction in Example 7 were:
[0129] temperature
time
16℃
60min
65℃
10min
4℃
∞
[0130] The ligation product was purified with a purification kit HiPure Nucleotide Remove Kit. The product was eluted with 25μl sterile ultrapure water, and 20μl was used in the subsequent steps.
[0131] (4) Asymmetrically amplify probes 3A and 3B obtained in step (3) to obtain a FISH probe library;
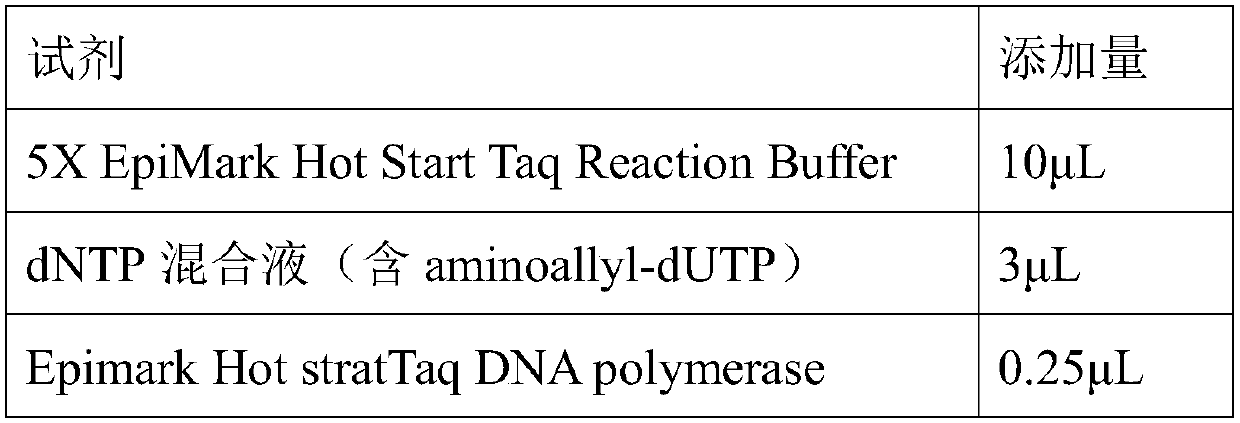
[0132] Asymmetric amplification reaction system:
[0133] Reagent
Add amount
5X EpiMark Hot Start Taq Reaction Buffer
10μL
dNTP mixture (containing aminoallyl-dUTP)
3μL
Epimark Hot stratTaq DNApolymerase
0.25μL
14.75μL
Primer F (1μM)
1μL
Primer R (20μM)
1μL
Probe 3A or 3B
20μL
[0134] Primer F and primer R are:
[0135] name
Base sequence (5’→3’)
Primer F
CCTCTGTATGCGCATCCTGTGAT
Primer R
TCGGAGACACTCAGAGATGTGATGG
[0136] In Examples 1-7 and 9, the dNTP mixture (containing aminoallyl-dUTP) is:
[0137] d...
Embodiment 1-9
[0147] Embodiments 1-9 are based on the basic embodiments and adjusted various parameters to obtain the following embodiments:
[0148]
[0149]
[0150]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com