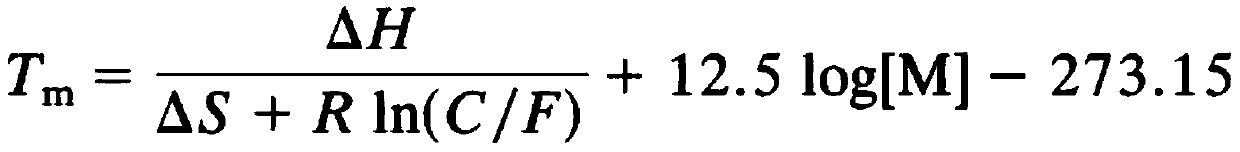Method for identifying four fusaria
A technology of Fusarium and Fusarium oxysporum, which is applied in biochemical equipment and methods, microbiological measurement/inspection, DNA/RNA fragments, etc., can solve the problems affecting the timeliness of food safety monitoring, the inability to identify pathogens in time, and the aggravation of patients' conditions and other problems, to achieve the effect of reducing the cost of experimental detection, low cost and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Embodiment 1 Differentiate the establishment and characteristic verification of four kinds of Fusarium methods
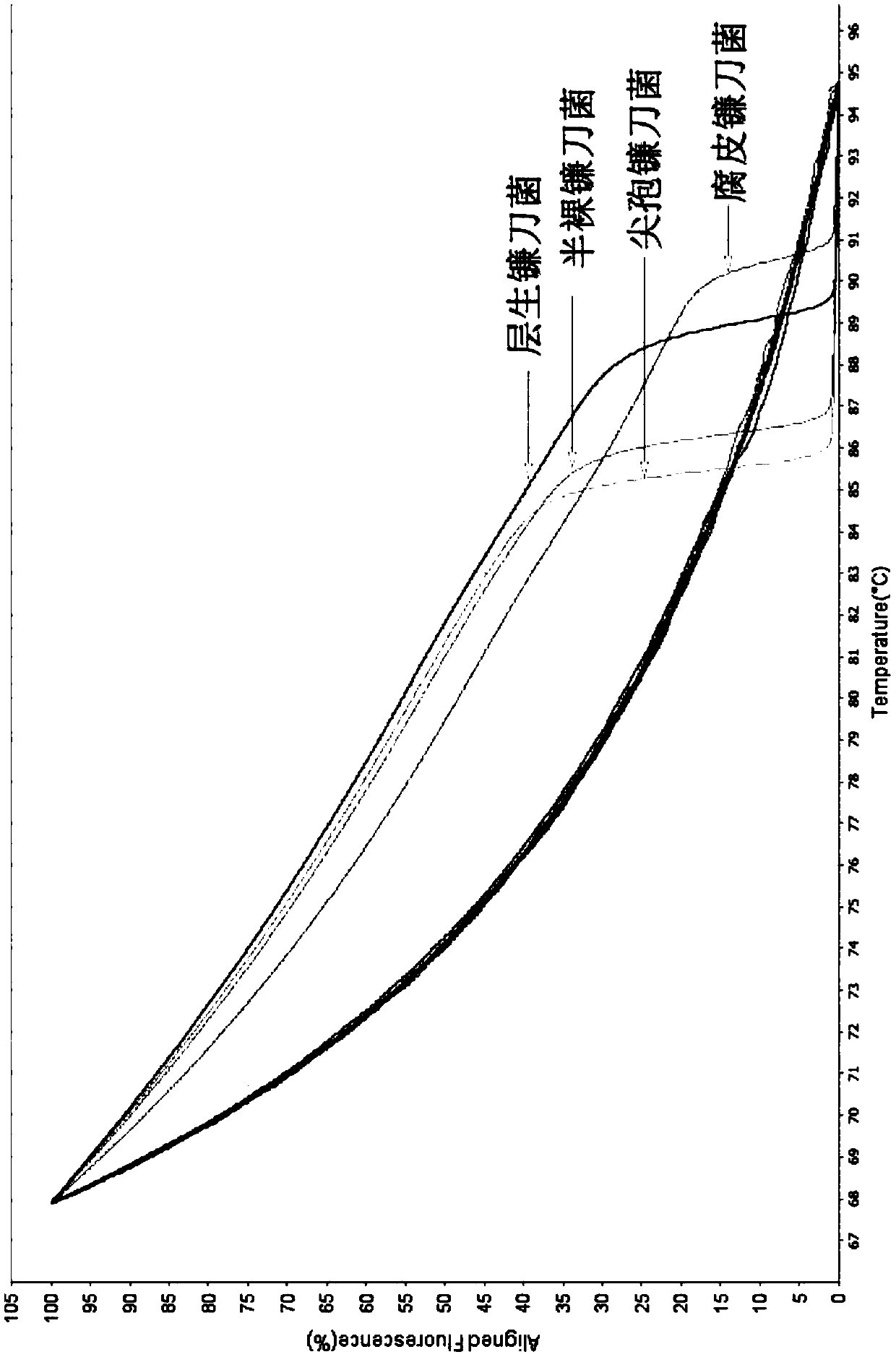
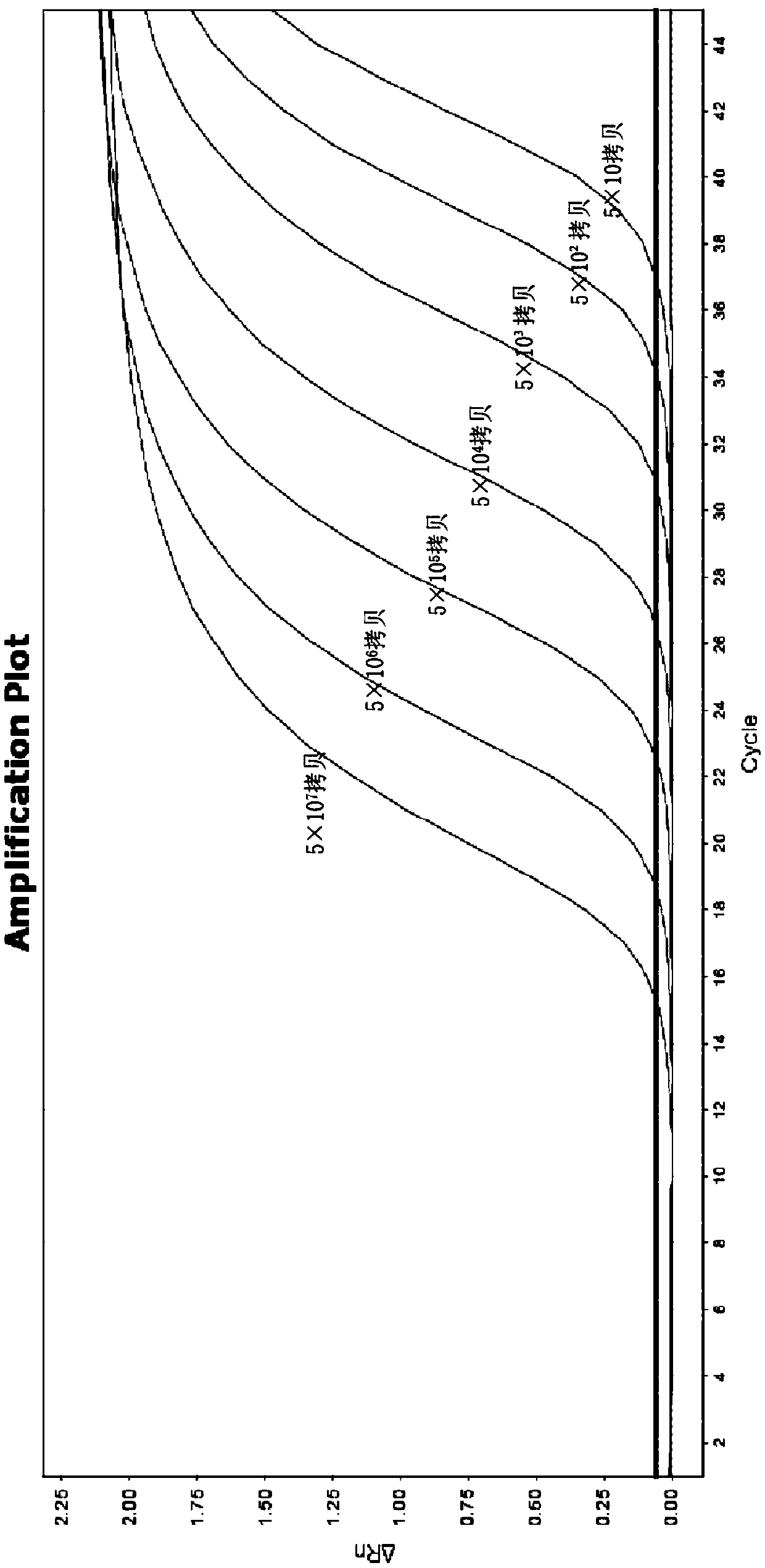
[0028] The present invention has carefully studied the genomic sequences of Fusarium proliferatum (Fusarium proliferatum), Fusarium oxysporum Schl., Fusarium semimitectum, and Fusarium solani. Look for differential target sequences in the ITS sequence, and design a pair of specific primers through careful design and screening, the nucleotide sequence of which is shown in SEQ ID NO.1-2, using this pair of primers can specifically identify the above-mentioned Four species of Fusarium. The design of this primer cannot be automatically obtained by computer software. It is necessary to select the target sequence fragment and length based on rich experience, and calculate the Tm value according to the following formula while considering the GC content of the sequence. After the primers were synthesized, trial and error was carried out to obtain a pair of optimal p...
Embodiment 2
[0046] Embodiment 2 Application of the inventive method
[0047] 1. Select 30 specimens of moldy corn kernels from Ningxia, inoculate them on SDA medium, culture them at 28°C and observe the growth of the colonies. After the growth of colonies, use the dilution coating plate method to select monoclonal colonies and culture them separately.
[0048] 2. The genomic DNA of the fungal sample to be tested was extracted by the rough extraction method, and the subsequent identification method was carried out with reference to the identification method in Example 1. The results were analyzed using ABI software, and the ITS differences of different Fusarium species were displayed based on the curve offset and curve shape changes through the standard curve. The identification results are shown in Table 2 below.
[0049] Table 2
[0050]
[0051]
[0052] For the samples for which the corresponding Fusarium was identified by the method in Example 1, the same traditional PCR meth...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com