A method for de novo synthesis of DNA shuffled libraries using chips to synthesize oligonucleotide libraries
An oligonucleotide library and library technology, applied in DNA preparation, recombinant DNA technology, chemical library, etc., to achieve the effect of reducing synthesis cost, increasing recombination rate, and increasing diversity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Design, synthesis and amplification of RFP oligo pool
[0038] 1. Design of RFP oligo pool
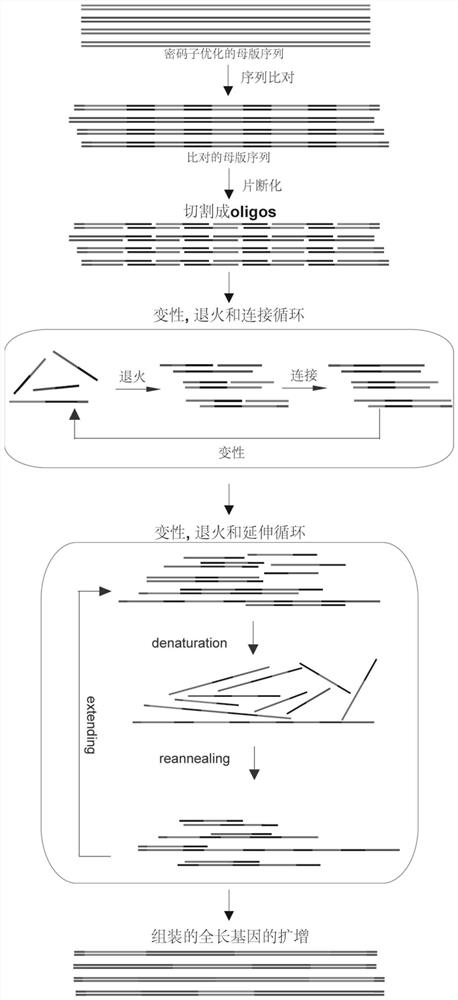
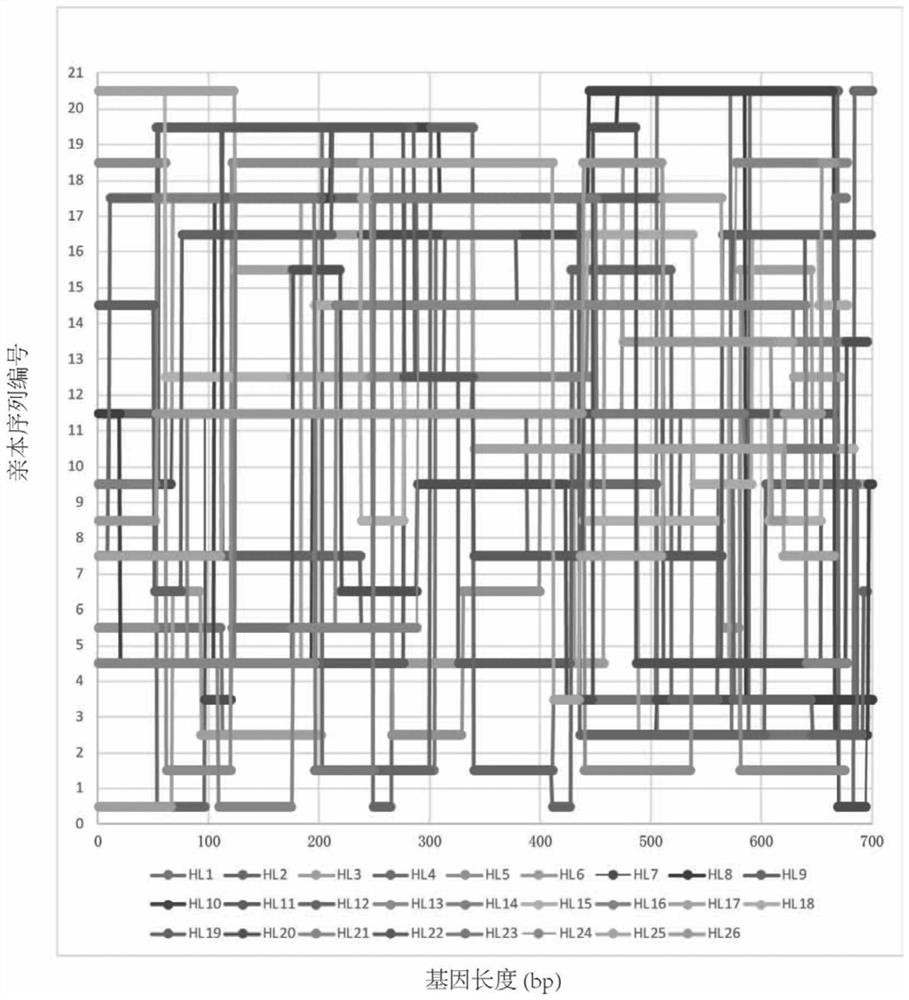
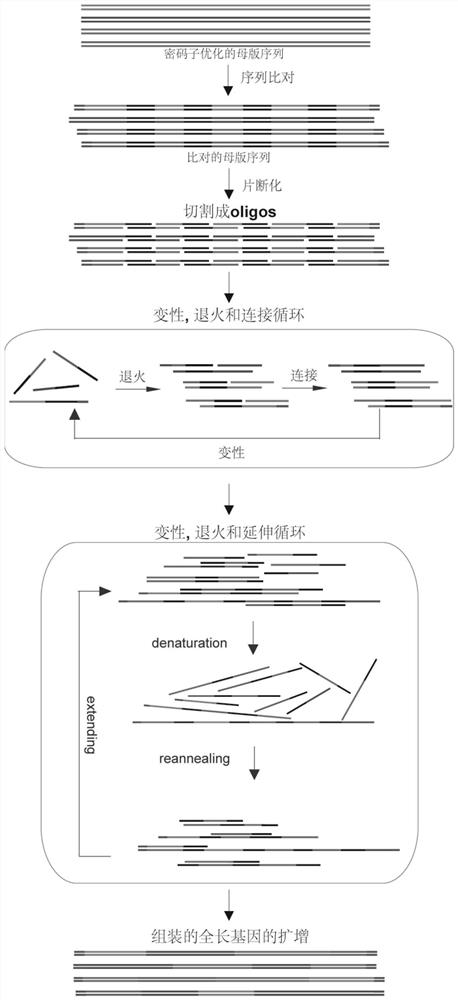
[0039] First, the 21 RFP parent sequences selected were codon-optimized according to the codon usage frequency of Escherichia coli, and the restriction endonuclease sites contained in the sequences were removed to form the optimized REP gene full-length sequence (SEQ ID No. .5-25). A pair of primer sequences are then added to the two segments of these sequences to facilitate subsequent amplification of the full-length sequence. These sequences are then compared, and the aligned sequences at the ends of their homologous regions are as follows figure 1 Interleaved cuts into multiple oligos were performed as shown. A pair of universal primers containing a type IIS restriction endonuclease Mly I site was added to each of the two segments of these oligos, which can be removed by enzymatic digestion before subsequent assembly reactions. These oligonucleotides can be ass...
Embodiment 2
[0056] Example 2: Analysis of Correction Efficiency of RFP oligo pool Synthesis Errors
[0057] In this implementation case, a round of error correction processing is performed on the RFP oligo pool described in Embodiment 1 by using MICC. The error-corrected sample is used as a template to re-amplify the ED-oligo pool by PCR, and then the oligo pool is assembled into a full-length recombinant library sequence by the method of LCR-PCA. Finally, these assembled full-length genes were cloned into the pMD18-T vector (Bao Bioengineering (Dalian) Co., Ltd.). Sequencing was performed, and the DNA analysis software Bioedit was used to compare the sequencing results with the full length of the parental genes.
[0058] The specific implementation steps are:
[0059] 1.MICC processing re-hybridized RFP oligo pool:
[0060] 1) Sample loading. Take 60 μl denatured and annealed RFP oligo pool, add 180 μl binding buffer and mix, the final concentration of DNA is 12.5ng / μl. Then load th...
Embodiment 3
[0110] Example 3, Plate induced expression of RFP recombinant library, positive clonal ratio analysis and positive clone sequencing analysis
[0111] 1. Construction of RFP recombinant library expression library
[0112] The full-length gene sequence of the recombinant library RFP-lib-NX prepared in Example 2 and the pET-21c vector were double digested with Nhe I and Xho I respectively, and the RFP-lib-NX sequence was inserted into the pET-21c vector by ligation , the plasmid pTaqMutS was obtained. The specific operation is as follows:
[0113] The amplified product RFP-lib-NX gene sequence and the pET-21c vector were digested with Nhe I and Xho I respectively, and ligated to construct the plasmid library "pet-RFP lib".
[0114] Enzyme digestion system of RFP-lib-NX gene sequence:
[0115]
[0116] Enzyme digestion system of pET-21c vector:
[0117]
[0118] Ligation system of RFP-lib-NX gene sequence and pET-21c vector:
[0119]
[0120]
[0121] The obtained...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com